HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays
- PMID: 15728728
- PMCID: PMC553304
- DOI: 10.1073/pnas.0407019102
HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays
Abstract
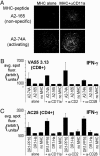
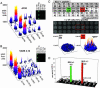
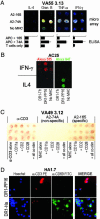
Identification of T cell epitopes is a vital but often slow and difficult step in studying the immune response to infectious agents and autoantigens. We report a spatially addressable technique for screening large numbers of T cell epitopes for both specific antigen recognition and functional activity induced. This system uses microarrays of immobilized, recombinant MHC-peptide complexes, costimulatory molecules, and cytokine-capture antibodies. The array elements act as synthetic antigen-presenting cells and specifically elicit T cell responses, including adhesion, secretion of cytokines, and modulation of surface markers. The method allows facile identification of pertinent T cell epitopes in a large number of candidates and simultaneous determination of the functional outcome of the interaction. Using this method, we have characterized the activation of human CD4(+) and CD8(+) T cells responding to vaccinia, influenza, HIV-1, and Epstein-Barr viruses.
Figures


References
-
- Collins, K. L. (2003) Curr. HIV Res. 1, 31-40. - PubMed
-
- Levitsky, V. & Masucci, M. G. (2002) Virus Res. 88, 71-86. - PubMed
-
- Terajima, M., Vapalahti, O., Van Epps, H. L., Vaheri, A. & Ennis, F. A. (2004) Microbes Infect. 6, 238-245. - PubMed
-
- Christen, U. & von Herrath, M. G. (2004) Mol. Immunol. 40, 1113-1120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

