Variation in the strength of selected codon usage bias among bacteria
- PMID: 15728743
- PMCID: PMC549432
- DOI: 10.1093/nar/gki242
Variation in the strength of selected codon usage bias among bacteria
Abstract
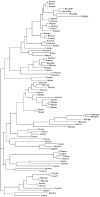
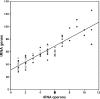
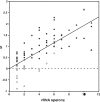
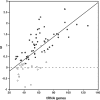
Among bacteria, many species have synonymous codon usage patterns that have been influenced by natural selection for those codons that are translated more accurately and/or efficiently. However, in other species selection appears to have been ineffective. Here, we introduce a population genetics-based model for quantifying the extent to which selection has been effective. The approach is applied to 80 phylogenetically diverse bacterial species for which whole genome sequences are available. The strength of selected codon usage bias, S, is found to vary substantially among species; in 30% of the genomes examined, there was no significant evidence that selection had been effective. Values of S are highly positively correlated with both the number of rRNA operons and the number of tRNA genes. These results are consistent with the hypothesis that species exposed to selection for rapid growth have more rRNA operons, more tRNA genes and more strongly selected codon usage bias. For example, Clostridium perfringens, the species with the highest value of S, can have a generation time as short as 7 min.
Figures



Similar articles
-
Translational Selection for Speed Is Not Sufficient to Explain Variation in Bacterial Codon Usage Bias.Genome Biol Evol. 2018 Feb 1;10(2):562-576. doi: 10.1093/gbe/evy018. Genome Biol Evol. 2018. PMID: 29385509 Free PMC article.
-
Codon Usage Optimization in the Prokaryotic Tree of Life: How Synonymous Codons Are Differentially Selected in Sequence Domains with Different Expression Levels and Degrees of Conservation.mBio. 2020 Jul 21;11(4):e00766-20. doi: 10.1128/mBio.00766-20. mBio. 2020. PMID: 32694138 Free PMC article.
-
Contributions of speed and accuracy to translational selection in bacteria.PLoS One. 2012;7(12):e51652. doi: 10.1371/journal.pone.0051652. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23272132 Free PMC article.
-
Forces that influence the evolution of codon bias.Philos Trans R Soc Lond B Biol Sci. 2010 Apr 27;365(1544):1203-12. doi: 10.1098/rstb.2009.0305. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20308095 Free PMC article. Review.
-
Codon Usage Bias: An Endless Tale.J Mol Evol. 2021 Dec;89(9-10):589-593. doi: 10.1007/s00239-021-10027-z. Epub 2021 Aug 12. J Mol Evol. 2021. PMID: 34383106 Review.
Cited by
-
Identification of Xenologs and Their Characteristic Low Expression Levels in the Cyanobacterium Synechococcus elongatus.J Mol Evol. 2015 Jun;80(5-6):292-304. doi: 10.1007/s00239-015-9684-x. Epub 2015 Jun 4. J Mol Evol. 2015. PMID: 26040248
-
Bacteria can maintain rRNA operons solely on plasmids for hundreds of millions of years.Nat Commun. 2023 Nov 14;14(1):7232. doi: 10.1038/s41467-023-42681-w. Nat Commun. 2023. PMID: 37963895 Free PMC article.
-
Pseudogenes act as a neutral reference for detecting selection in prokaryotic pangenomes.Nat Ecol Evol. 2024 Feb;8(2):304-314. doi: 10.1038/s41559-023-02268-6. Epub 2024 Jan 4. Nat Ecol Evol. 2024. PMID: 38177690
-
Depletion of Shine-Dalgarno Sequences Within Bacterial Coding Regions Is Expression Dependent.G3 (Bethesda). 2016 Nov 8;6(11):3467-3474. doi: 10.1534/g3.116.032227. G3 (Bethesda). 2016. PMID: 27605518 Free PMC article.
-
Transfer RNA gene numbers may not be completely responsible for the codon usage bias in asparagine, isoleucine, phenylalanine, and tyrosine in the high expression genes in bacteria.J Mol Evol. 2012 Aug;75(1-2):34-42. doi: 10.1007/s00239-012-9524-1. Epub 2012 Oct 2. J Mol Evol. 2012. PMID: 23053196
References
-
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. - PubMed
-
- Sharp P.M., Cowe E., Higgins D.G., Shields D.C., Wolfe K.H., Wright F. Codon usage in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. - PMC - PubMed
-
- Sharp P.M., Li W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

