Functional cartography of complex metabolic networks
- PMID: 15729348
- PMCID: PMC2175124
- DOI: 10.1038/nature03288
Functional cartography of complex metabolic networks
Abstract
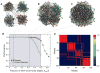
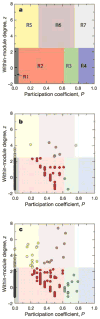
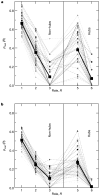
High-throughput techniques are leading to an explosive growth in the size of biological databases and creating the opportunity to revolutionize our understanding of life and disease. Interpretation of these data remains, however, a major scientific challenge. Here, we propose a methodology that enables us to extract and display information contained in complex networks. Specifically, we demonstrate that we can find functional modules in complex networks, and classify nodes into universal roles according to their pattern of intra- and inter-module connections. The method thus yields a 'cartographic representation' of complex networks. Metabolic networks are among the most challenging biological networks and, arguably, the ones with most potential for immediate applicability. We use our method to analyse the metabolic networks of twelve organisms from three different superkingdoms. We find that, typically, 80% of the nodes are only connected to other nodes within their respective modules, and that nodes with different roles are affected by different evolutionary constraints and pressures. Remarkably, we find that metabolites that participate in only a few reactions but that connect different modules are more conserved than hubs whose links are mostly within a single module.
Conflict of interest statement
Figures

References
-
- Albert R, Barabási AL. Statistical mechanics of complex networks. Rev Mod Phys. 2002;74:47–97.
-
- Amaral LAN, Ottino J. Complex networks: Augmenting the framework for the study of complex systems. Eur Phys J B. 2004;38:147–162.
-
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular biology. Nature. 1999;402(Suppl):C47–C52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

