Sialic Acid metabolism and systemic pasteurellosis
- PMID: 15731025
- PMCID: PMC1064920
- DOI: 10.1128/IAI.73.3.1284-1294.2005
Sialic Acid metabolism and systemic pasteurellosis
Abstract
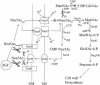
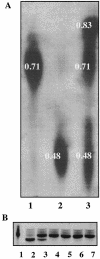
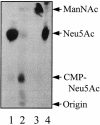
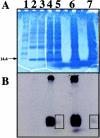
Pasteurella multocida subsp. multocida is a commensal and opportunistic pathogen of food animals, wildlife, and pets and a zoonotic cause of human infection arising from contacts with these animals. Here, an investigation of multiple serotype A strains demonstrated the occurrence of membrane sialyltransferase. Although P. multocida lacks the genes for the two earliest steps in de novo sialic acid synthesis, adding sialic acid to the growth medium resulted in uptake, activation, and subsequent transfer of sialic acid to a membrane acceptor resembling lipooligosaccharide. Two candidate-activating enzymes with homology to Escherichia coli cytidine 5'-monophospho-N-acetylneuraminate synthetase were overproduced as histidine-tagged polypeptides. The synthetase encoded by pm0187 was at least 37 times more active than the pm1710 gene product, suggesting pm0187 encodes the primary sialic acid cytidylyltransferase in P. multocida. A sialate aldolase (pm1715) mutant unable to initiate dissimilation of internalized sialic acid was not attenuated in the CD-1 mouse model of systemic pasteurellosis, indicating that the nutritional function of sialate catabolism is not required for systemic disease. In contrast, the attenuation of a sialate uptake-deficient mutant supports the essential role in pathogenesis of a sialylation mechanism that is dependent on an environmental (host) supply of sialic acid. The combined results provide the first direct evidence of sialylation by a precursor scavenging mechanism in pasteurellae and of a potential tripartite ATP-independent periplasmic sialate transporter in any species.
Figures




Similar articles
-
Sialic acid uptake is necessary for virulence of Pasteurella multocida in turkeys.Microb Pathog. 2009 Jun;46(6):337-44. doi: 10.1016/j.micpath.2009.04.003. Epub 2009 Apr 12. Microb Pathog. 2009. PMID: 19366625
-
Genetic engineering of Escherichia coli for the economical production of sialylated oligosaccharides.J Biotechnol. 2008 Apr 30;134(3-4):261-5. doi: 10.1016/j.jbiotec.2008.02.010. Epub 2008 Mar 10. J Biotechnol. 2008. PMID: 18378033
-
Modulating the regioselectivity of a Pasteurella multocida sialyltransferase for biocatalytic production of 3'- and 6'-sialyllactose.Enzyme Microb Technol. 2015 Oct;78:54-62. doi: 10.1016/j.enzmictec.2015.06.012. Epub 2015 Jun 20. Enzyme Microb Technol. 2015. PMID: 26215345
-
Sialic acid activation.Glycobiology. 1991 Nov;1(5):441-7. doi: 10.1093/glycob/1.5.441. Glycobiology. 1991. PMID: 1668304 Review.
-
Pasteurella multocida and bovine respiratory disease.Anim Health Res Rev. 2007 Dec;8(2):129-50. doi: 10.1017/S1466252307001399. Anim Health Res Rev. 2007. PMID: 18218157 Review.
Cited by
-
Catabolism of N-acetylneuraminic acid, a fitness function of the food-borne lactic acid bacterium Lactobacillus sakei, involves two newly characterized proteins.Appl Environ Microbiol. 2013 Mar;79(6):2012-8. doi: 10.1128/AEM.03301-12. Epub 2013 Jan 18. Appl Environ Microbiol. 2013. PMID: 23335758 Free PMC article.
-
Structural and Biosynthetic Diversity of Nonulosonic Acids (NulOs) That Decorate Surface Structures in Bacteria.Trends Microbiol. 2021 Feb;29(2):142-157. doi: 10.1016/j.tim.2020.08.002. Epub 2020 Sep 17. Trends Microbiol. 2021. PMID: 32950378 Free PMC article. Review.
-
Pasteurella multocida: Genotypes and Genomics.Microbiol Mol Biol Rev. 2019 Sep 4;83(4):e00014-19. doi: 10.1128/MMBR.00014-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31484691 Free PMC article. Review.
-
Sialic acid mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence.BMC Microbiol. 2010 Feb 16;10:48. doi: 10.1186/1471-2180-10-48. BMC Microbiol. 2010. PMID: 20158882 Free PMC article.
-
PmST2: a novel Pasteurella multocida glycolipid α2-3-sialyltransferase.Glycobiology. 2011 Sep;21(9):1206-16. doi: 10.1093/glycob/cwr054. Epub 2011 Apr 21. Glycobiology. 2011. PMID: 21515586 Free PMC article.
References
-
- Adlam, C., and J. M. Rutter. 1989. Pasteurella and pasteurellosis. Academic Press, Ltd., London, England.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Angata, T., and A. Varki. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439-469. - PubMed
-
- Berge, A., A. Fagergren, and S. H. Stierenstedt. 2002. Pasteurella multocida septicemia in 2 Swedish patients. Scand. J. Infect. Dis. 34:138-139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

