The fimYZ genes regulate Salmonella enterica Serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility
- PMID: 15731035
- PMCID: PMC1064959
- DOI: 10.1128/IAI.73.3.1377-1385.2005
The fimYZ genes regulate Salmonella enterica Serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility
Abstract
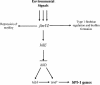
An important step in Salmonella enterica serovar Typhimurium virulence is the ability to invade the intestinal epithelium. The invasion process requires a large number of genes encoded on Salmonella pathogenicity island 1 (SPI-1) at centisome 63 as well as genes located in other positions throughout the chromosome. Expression of the invasive phenotype is tightly regulated by environmental cues that are processed by a complex regulatory scheme. A central player in the invasion regulatory pathway is the HilA protein, which is transcriptional activator belonging to the OmpR/ToxR family. A number of positive regulators (hilC, hilD, fis, sirA/barA, csrAB, phoBR, fadD, envZ/ompR, and fliZ) and negative regulators (hha, hilE, lon, ams, phoPc and pag) have been identified that are able to alter expression of hilA transcription. Recent work has found that hilA transcription requires the HilD protein for activation. Other work has emphasized the importance of HilE as a negative regulator of hilA. Overexpression of hilE superrepresses hilA transcription, as well as the invasive phenotype. Two-hybrid experiments suggest that HilE exerts its regulatory influence on hilA through protein-protein interactions with HilD as the protein does not bind to the hilA promoter nor does it affect hilD transcription. As it seems likely that hilE plays an important role in translating environmental signals into invasion gene regulation, we have attempted to identify how the hilE gene itself is regulated. Our results indicate that the fimYZ genes, response regulatory proteins involved in type 1 fimbrial gene expression and recently implicated in motility gene regulation, are important activators of hilE expression. These findings indicate that invasion gene expression is coregulated with motility and adherence and provide experimental evidence that the expression of these virulence phenotypes is a subset of the overall regulation of bacterial physiology.
Figures




Similar articles
-
HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype.Infect Immun. 2003 Mar;71(3):1295-305. doi: 10.1128/IAI.71.3.1295-1305.2003. Infect Immun. 2003. PMID: 12595445 Free PMC article.
-
Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators.J Bacteriol. 2003 Jan;185(2):525-33. doi: 10.1128/JB.185.2.525-533.2003. J Bacteriol. 2003. PMID: 12511499 Free PMC article.
-
Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium.J Bacteriol. 2001 May;183(9):2733-45. doi: 10.1128/JB.183.9.2733-2745.2001. J Bacteriol. 2001. PMID: 11292791 Free PMC article.
-
Salmonella invasion gene regulation: a story of environmental awareness.J Microbiol. 2005 Feb;43 Spec No:110-7. J Microbiol. 2005. PMID: 15765064 Review.
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
Cited by
-
HilE Regulates HilD by Blocking DNA Binding in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2018 Mar 26;200(8):e00750-17. doi: 10.1128/JB.00750-17. Print 2018 Apr 15. J Bacteriol. 2018. PMID: 29378886 Free PMC article.
-
AraC-type regulators HilC and RtsA are directly controlled by an intestinal fatty acid to regulate Salmonella invasion.Mol Microbiol. 2021 Dec;116(6):1464-1475. doi: 10.1111/mmi.14835. Epub 2021 Nov 8. Mol Microbiol. 2021. PMID: 34687258 Free PMC article.
-
Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease.J Bacteriol. 2008 Apr;190(7):2470-8. doi: 10.1128/JB.01385-07. Epub 2008 Feb 1. J Bacteriol. 2008. PMID: 18245288 Free PMC article.
-
Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium.Infect Immun. 2015 Mar;83(3):978-85. doi: 10.1128/IAI.02506-14. Epub 2014 Dec 29. Infect Immun. 2015. PMID: 25547794 Free PMC article.
-
Role of FimW, FimY, and FimZ in regulating the expression of type i fimbriae in Salmonella enterica serovar Typhimurium.J Bacteriol. 2009 May;191(9):3003-10. doi: 10.1128/JB.01694-08. Epub 2009 Feb 13. J Bacteriol. 2009. PMID: 19218381 Free PMC article.
References
-
- Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-728. - PubMed
-
- Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:1872-1882. - PubMed
-
- Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. - PubMed
-
- Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

