Object-oriented biological system integration: a SARS coronavirus example
- PMID: 15731211
- PMCID: PMC7197705
- DOI: 10.1093/bioinformatics/bti344
Object-oriented biological system integration: a SARS coronavirus example
Abstract
Motivation: The importance of studying biology at the system level has been well recognized, yet there is no well-defined process or consistent methodology to integrate and represent biological information at this level. To overcome this hurdle, a blending of disciplines such as computer science and biology is necessary.
Results: By applying an adapted, sequential software engineering process, a complex biological system (severe acquired respiratory syndrome-coronavirus viral infection) has been reverse-engineered and represented as an object-oriented software system. The scalability of this object-oriented software engineering approach indicates that we can apply this technology for the integration of large complex biological systems.
Availability: A navigable web-based version of the system is freely available at http://people.musc.edu/~zhengw/SARS/Software-Process.htm
Figures
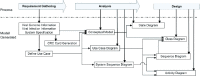

 ), representing extensions of the objects below. Functions and events associated with these bioentities are modeled as messages sent to the corresponding objects. These message calls, indicated by solid arrows, are made between objects via connection of their lifelines. Messages contain a message name followed by parameters, which must be passed into the object and/or outputted from the object, e.g. translation machinery executes its function of translating viral mRNA by receiving a translate message ‘translate (in: Positive_Genomic_RNA)’ from the cell. This message accepts a Positive_Genomic_RNA as a template, indicated by ‘in: Positive_Genomic_RNA’, then a ‘Viral_Replicase’ is created and returned. These return values are displayed below the message call as a named dotted arrow extending in the opposite direction as the original message call. Where return values are not explicitly indicated, as in an object calling itself (
), representing extensions of the objects below. Functions and events associated with these bioentities are modeled as messages sent to the corresponding objects. These message calls, indicated by solid arrows, are made between objects via connection of their lifelines. Messages contain a message name followed by parameters, which must be passed into the object and/or outputted from the object, e.g. translation machinery executes its function of translating viral mRNA by receiving a translate message ‘translate (in: Positive_Genomic_RNA)’ from the cell. This message accepts a Positive_Genomic_RNA as a template, indicated by ‘in: Positive_Genomic_RNA’, then a ‘Viral_Replicase’ is created and returned. These return values are displayed below the message call as a named dotted arrow extending in the opposite direction as the original message call. Where return values are not explicitly indicated, as in an object calling itself ( ), an ‘out’ followed by a return value is substituted. Events in gray boxes are shown in more detail in Supplemental Figures 7–9.
), an ‘out’ followed by a return value is substituted. Events in gray boxes are shown in more detail in Supplemental Figures 7–9.
 ) and pseudo-end (
) and pseudo-end ( ) states. Action states, represented by rounded rectangles (
) states. Action states, represented by rounded rectangles ( ) are captured during different time periods. This diagram can represent sequential as well as concurrent events. Concurrent events are represented by forks (
) are captured during different time periods. This diagram can represent sequential as well as concurrent events. Concurrent events are represented by forks ( ) and merges (
) and merges ( ). This architecture signifies that multiple events are possible simultaneously and the occurrence of at least one of these events is enough to continue the flow of control. In contrast, the flow of control may also be conditional, e.g. decision diamonds (⋄) represent a point at which alternative events can occur. At these decision diamonds, guard conditions must be satisfied for the flow of control to continue, or else the flow of control follows an alternative route.
). This architecture signifies that multiple events are possible simultaneously and the occurrence of at least one of these events is enough to continue the flow of control. In contrast, the flow of control may also be conditional, e.g. decision diamonds (⋄) represent a point at which alternative events can occur. At these decision diamonds, guard conditions must be satisfied for the flow of control to continue, or else the flow of control follows an alternative route.
 ) at the end indicate composition. These are read from the diamond end, e.g. as Virus contains Structural_Proteins. Lines with open triangles (
) at the end indicate composition. These are read from the diamond end, e.g. as Virus contains Structural_Proteins. Lines with open triangles ( ) represent generalizations. These are read from the triangle end, as Viral_Protein is a general type of Structural_Protein. Other interactions are represented as named, binary associations (
) represent generalizations. These are read from the triangle end, as Viral_Protein is a general type of Structural_Protein. Other interactions are represented as named, binary associations ( ). Interactions may contain cardinalities at their ends indicating the number of objects that interact with another object. As an example from above, one virus can infect one cell and one cell can be infected by one to many viruses. Grayed objects and interactions are shown in more detail in Supplemental Figures 14–16. Viral components (orange); cellular components (blue).
). Interactions may contain cardinalities at their ends indicating the number of objects that interact with another object. As an example from above, one virus can infect one cell and one cell can be infected by one to many viruses. Grayed objects and interactions are shown in more detail in Supplemental Figures 14–16. Viral components (orange); cellular components (blue).Similar articles
-
Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication.J Virol. 2009 Mar;83(5):2368-73. doi: 10.1128/JVI.02371-08. Epub 2008 Dec 17. J Virol. 2009. PMID: 19091867 Free PMC article.
-
Hosting the severe acute respiratory syndrome coronavirus: specific cell factors required for infection.Cell Microbiol. 2006 Aug;8(8):1211-8. doi: 10.1111/j.1462-5822.2006.00744.x. Epub 2006 Jun 27. Cell Microbiol. 2006. PMID: 16803585 Free PMC article. Review.
-
SARS coronavirus: a new challenge for prevention and therapy.J Clin Invest. 2003 Jun;111(11):1605-9. doi: 10.1172/JCI18819. J Clin Invest. 2003. PMID: 12782660 Free PMC article. Review. No abstract available.
-
Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence.Trends Microbiol. 2007 Feb;15(2):51-3. doi: 10.1016/j.tim.2006.12.005. Epub 2007 Jan 4. Trends Microbiol. 2007. PMID: 17207625 Free PMC article. Review.
-
Molecular pathogenesis of severe acute respiratory syndrome.Microbes Infect. 2007 Jan;9(1):119-26. doi: 10.1016/j.micinf.2006.06.012. Epub 2006 Sep 28. Microbes Infect. 2007. PMID: 17142081 Free PMC article. Review.
Cited by
-
Incorporating Time Delays in the Mathematical Modelling of the Human Immune Response in Viral Infections.Procedia Comput Sci. 2021;185:144-151. doi: 10.1016/j.procs.2021.05.016. Epub 2021 Jun 10. Procedia Comput Sci. 2021. PMID: 34131452 Free PMC article.
-
A rigorous approach to facilitate and guarantee the correctness of the genetic testing management in human genome information systems.BMC Genomics. 2011 Dec 22;12 Suppl 4(Suppl 4):S13. doi: 10.1186/1471-2164-12-S4-S13. Epub 2011 Dec 22. BMC Genomics. 2011. PMID: 22369688 Free PMC article.
-
Integration of the Gene Ontology into an object-oriented architecture.BMC Bioinformatics. 2005 May 10;6:113. doi: 10.1186/1471-2105-6-113. BMC Bioinformatics. 2005. PMID: 15885145 Free PMC article.
-
Genome3D: a viewer-model framework for integrating and visualizing multi-scale epigenomic information within a three-dimensional genome.BMC Bioinformatics. 2010 Sep 2;11:444. doi: 10.1186/1471-2105-11-444. BMC Bioinformatics. 2010. PMID: 20813045 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

