Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome
- PMID: 15731256
- PMCID: PMC1075693
- DOI: 10.1128/JVI.79.6.3615-3626.2005
Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome
Abstract
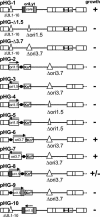
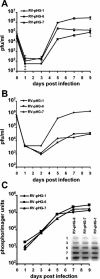
During the lytic phase of infection, replication of herpesvirus genomes initiates at the lytic origin of replication, oriLyt. Many herpesviruses harbor more than one lytic origin, but so far, only one oriLyt has been identified for human cytomegalovirus (HCMV). Evidence for the existence of additional lytic origins of HCMV has remained elusive. On the basis of transient replication assays with cloned viral fragments, HCMV oriLyt was described as a core region of 1.5 kbp (minimal oriLyt) flanked by auxiliary sequences required for maximal replication activity (complete oriLyt). It remained unclear whether minimal oriLyt alone can drive the replication of HCMV in the absence of its accessory regions. To investigate the sequence requirements of oriLyt in the context of the viral genome, mutant genomes were constructed lacking either minimal or complete oriLyt. These genomes were not infectious, suggesting that HCMV contains only one lytic origin of replication. Either minimal or complete oriLyt was then ectopically reinserted into the oriLyt-depleted genomes. Only the mutant genomes carrying complete oriLyt led to infectious progeny. Remarkably, inversion of the 1.5-kbp core origin relative to its flanking regions resulted in a replication-defective genome. Mutant genomes carrying minimal oriLyt plus the left flanking region gave rise to minifoci, but genomes harboring minimal oriLyt together with the right flanking region were noninfectious. We conclude that the previously defined minimal lytic origin is not sufficient to drive replication of the HCMV genome. Rather, our results underline the importance of the accessory regions and their correct arrangement for the function of HCMV oriLyt.
Figures
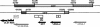





Similar articles
-
Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays.J Virol. 1996 Nov;70(11):7398-413. doi: 10.1128/JVI.70.11.7398-7413.1996. J Virol. 1996. PMID: 8892858 Free PMC article.
-
Human cytomegalovirus oriLyt sequence requirements.J Virol. 1998 Jun;72(6):4989-96. doi: 10.1128/JVI.72.6.4989-4996.1998. J Virol. 1998. PMID: 9573268 Free PMC article.
-
Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication.J Virol. 1992 Jun;66(6):3373-84. doi: 10.1128/JVI.66.6.3373-3384.1992. J Virol. 1992. PMID: 1316454 Free PMC article.
-
[The mechanisms of human cytomegalovirus DNA replication].Nihon Rinsho. 1998 Jan;56(1):36-43. Nihon Rinsho. 1998. PMID: 9465662 Review. Japanese.
-
Nuts and bolts of human cytomegalovirus lytic DNA replication.Curr Top Microbiol Immunol. 2008;325:153-66. doi: 10.1007/978-3-540-77349-8_9. Curr Top Microbiol Immunol. 2008. PMID: 18637505 Review.
Cited by
-
A novel strain of cynomolgus macaque cytomegalovirus: implications for host-virus co-evolution.BMC Genomics. 2016 Apr 5;17:277. doi: 10.1186/s12864-016-2588-3. BMC Genomics. 2016. PMID: 27044312 Free PMC article.
-
Host-viral effects of chromatin assembly factor 1 interaction with HCMV IE2.Cell Res. 2011 Aug;21(8):1230-47. doi: 10.1038/cr.2011.53. Epub 2011 Mar 29. Cell Res. 2011. PMID: 21445097 Free PMC article.
-
Regulation of the transcription and replication cycle of human cytomegalovirus is insensitive to genetic elimination of the cognate NF-kappaB binding sites in the enhancer.J Virol. 2006 Oct;80(19):9899-904. doi: 10.1128/JVI.00640-06. J Virol. 2006. PMID: 16973595 Free PMC article.
-
A peptide inhibitor of cytomegalovirus infection from human hemofiltrate.Antimicrob Agents Chemother. 2013 Oct;57(10):4751-60. doi: 10.1128/AAC.00854-13. Epub 2013 Jul 15. Antimicrob Agents Chemother. 2013. PMID: 23856778 Free PMC article.
-
Labyrinthopeptins Exert Broad-Spectrum Antiviral Activity through Lipid-Binding-Mediated Virolysis.J Virol. 2020 Jan 6;94(2):e01471-19. doi: 10.1128/JVI.01471-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666384 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

