Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production
- PMID: 15731257
- PMCID: PMC1075700
- DOI: 10.1128/JVI.79.6.3627-3638.2005
Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production
Abstract
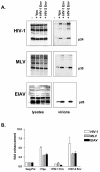
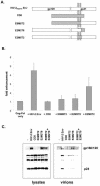
Primate lentiviruses code for a protein that stimulates virus production. In human immunodeficiency virus type 1 (HIV-1), the activity is provided by the accessory protein, Vpu, while in HIV-2 and simian immunodeficiency virus it is a property of the envelope (Env) glycoprotein. Using a group of diverse retroviruses and cell types, we have confirmed the functional equivalence of the two proteins. However, despite these similarities, the two proteins have markedly different functional domains. While the Vpu activity is associated primarily with its membrane-spanning region, we have determined that the HIV-2 Env activity requires both the cytoplasmic tail and ectodomain of the protein, with the membrane-spanning domain being less important. Within the Env cytoplasmic tail, we further defined the necessary sequence as a membrane-proximal tyrosine-based motif. Providing the two Env regions separately as distinct CD8 chimeric proteins did not increase virus release. This suggests that the two domains must be either contained within a single protein or closely associated within a multiprotein oligomer, such as the Env trimer, in order to function. Finally, we observed that wild-type levels of incorporation of the HIV-2 Env into budding viruses were not required for this activity.
Figures





References
-
- Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. - PubMed
-
- Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. - PMC - PubMed
-
- Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

