Lambda interferon inhibits hepatitis B and C virus replication
- PMID: 15731279
- PMCID: PMC1075734
- DOI: 10.1128/JVI.79.6.3851-3854.2005
Lambda interferon inhibits hepatitis B and C virus replication
Abstract
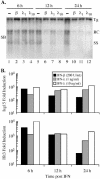
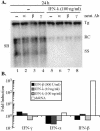
Lambda interferon (IFN-lambda) induces an intracellular IFN-alpha/beta-like antiviral response through a receptor complex distinct from the IFN-alpha/beta receptor. We therefore determined the ability of IFN-lambda to inhibit hepatitis B virus (HBV) and hepatitis C virus (HCV) replication. IFN-lambda inhibits HBV replication in a differentiated murine hepatocyte cell line with kinetics and efficiency similar to IFN-alpha/beta and does not require the expression of IFN-alpha/beta or IFN-gamma. Furthermore, IFN-lambda blocked the replication of a subgenomic and a full-length genomic HCV replicon in human hepatocyte Huh7 cells. These results suggest the possibility that IFN-lambda may be therapeutically useful in the treatment of chronic HBV or HCV infection.
Figures
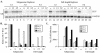
Similar articles
-
ME3738 enhances the effect of interferon and inhibits hepatitis C virus replication both in vitro and in vivo.J Hepatol. 2011 Jul;55(1):11-8. doi: 10.1016/j.jhep.2010.10.017. Epub 2010 Nov 29. J Hepatol. 2011. PMID: 21145867
-
HBV Bypasses the Innate Immune Response and Does Not Protect HCV From Antiviral Activity of Interferon.Gastroenterology. 2018 May;154(6):1791-1804.e22. doi: 10.1053/j.gastro.2018.01.044. Epub 2018 Feb 1. Gastroenterology. 2018. PMID: 29410097
-
Hepatitis C virus replication is inhibited by 22beta-methoxyolean-12-ene-3beta, 24(4beta)-diol (ME3738) through enhancing interferon-beta.Hepatology. 2008 Jul;48(1):59-69. doi: 10.1002/hep.22289. Hepatology. 2008. PMID: 18459156
-
[The present state of research in direct antiviral mechanism of interferon on hepatitis B virus].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2009 Dec;26(6):1358-62, 1371. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2009. PMID: 20095503 Review. Chinese.
-
[Interferon].Nihon Rinsho. 2012 Apr;70(4):620-4. Nihon Rinsho. 2012. PMID: 22568143 Review. Japanese.
Cited by
-
Dendritic cells in hepatitis C virus infection: key players in the IFNL3-genotype response.World J Gastroenterol. 2014 Dec 21;20(47):17830-8. doi: 10.3748/wjg.v20.i47.17830. World J Gastroenterol. 2014. PMID: 25548481 Free PMC article. Review.
-
Dysregulation of retinoic acid receptor diminishes hepatocyte permissiveness to hepatitis B virus infection through modulation of sodium taurocholate cotransporting polypeptide (NTCP) expression.J Biol Chem. 2015 Feb 27;290(9):5673-84. doi: 10.1074/jbc.M114.602540. Epub 2014 Dec 30. J Biol Chem. 2015. PMID: 25550158 Free PMC article.
-
EPSTI1 Is Involved in IL-28A-Mediated Inhibition of HCV Infection.Mediators Inflamm. 2015;2015:716315. doi: 10.1155/2015/716315. Epub 2015 Jun 3. Mediators Inflamm. 2015. PMID: 26146465 Free PMC article.
-
Gene Expression and Antiviral Activity of Interleukin-35 in Response to Influenza A Virus Infection.J Biol Chem. 2016 Aug 5;291(32):16863-76. doi: 10.1074/jbc.M115.693101. Epub 2016 Jun 15. J Biol Chem. 2016. PMID: 27307042 Free PMC article.
-
Meta-analysis of associations of interleukin-28B polymorphisms rs8099917 and rs12979860 with development of hepatitis virus-related hepatocellular carcinoma.Onco Targets Ther. 2016 May 30;9:3249-57. doi: 10.2147/OTT.S104904. eCollection 2016. Onco Targets Ther. 2016. PMID: 27313470 Free PMC article. Review.
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Coccia, E. M., M. Severa, E. Giacomini, D. Monneron, M. E. Remoli, I. Julkunen, M. Cella, R. Lande, and G. Uze. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34:796-805. - PubMed
-
- Donnelly, R. P., F. Sheikh, S. V. Kotenko, and H. Dickensheets. 2004. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 76:314-321. - PubMed
-
- Farrell, P. J., R. J. Broeze, and P. Lengyel. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279:523-525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

