Selection of functional human antibodies from retroviral display libraries
- PMID: 15731328
- PMCID: PMC549574
- DOI: 10.1093/nar/gni033
Selection of functional human antibodies from retroviral display libraries
Abstract
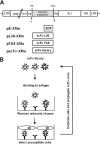
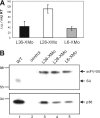
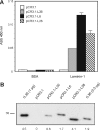
Antibody library technology represents a powerful tool for the discovery and design of antibodies with high affinity and specificity for their targets. To extend the technique to the expression and selection of antibody libraries in an eukaryotic environment, we provide here a proof of concept that retroviruses can be engineered for the display and selection of variable single-chain fragment (scFv) libraries. A retroviral library displaying the repertoire obtained after a single round of selection of a human synthetic scFv phage display library on laminin was generated. For selection, antigen-bound virus was efficiently recovered by an overlay with cells permissive for infection. This approach allowed more than 10(3)-fold enrichment of antigen binders in a single selection cycle. After three selection cycles, several scFvs were recovered showing similar laminin-binding activities but improved expression levels in mammalian cells as compared with a laminin-specific scFv selected by the conventional phage display approach. Thus, translational problems that occur when phage-selected antibodies have to be transferred onto mammalian expression systems to exert their therapeutic potential can be avoided by the use of retroviral display libraries.
Figures



References
-
- Wittrup K.D. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 2001;12:395–399. - PubMed
-
- Colas P. Combinatorial protein reagents to manipulate protein function. Curr. Opin. Chem. Biol. 2000;4:54–59. - PubMed
-
- Hoogenboom H.R., de Bruine A.P., Hufton S.E., Hoet R.M., Arends J.W., Roovers R.C. Antibody phage display technology and its applications. Immunotechnology. 1998;4:1–20. - PubMed
-
- Sanz L., Kristensen P., Blanco B., Facteau S., Russell S.J., Winter G., Alvarez-Vallina L. Single-chain antibody-based gene therapy: inhibition of tumor growth by in situ production of phage-derived human antibody fragments blocking functionally active sites of cell-associated matrices. Gene Ther. 2002;9:1049–1053. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

