Multiplexed discovery of sequence polymorphisms using base-specific cleavage and MALDI-TOF MS
- PMID: 15731331
- PMCID: PMC549577
- DOI: 10.1093/nar/gni038
Multiplexed discovery of sequence polymorphisms using base-specific cleavage and MALDI-TOF MS
Abstract
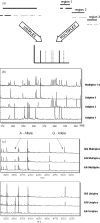
The completion of the Human Genome Project provides researchers with a reference sequence that covers about 99% of the gene-containing regions and is more than 99.9% accurate. Sequence drafts and completed sequences for several other species are also available to researchers worldwide. The ongoing effort to provide more and more genomic reference information now enables the detection of deviations from this 'genetic blueprint'. Comparative sequencing projects will play a major role in elucidating the meaning of the genetic code and in establishing a correlation between genotype and phenotype. As part of this effort, a number of projects will focus on distinct functional aspects, like resequencing of exons or HLA determining regions. Typically these target regions are short in length and their analysis does not require long read length. To find an efficient solution for these applications, we developed a novel method that allows simultaneous analysis of multiple independent target regions (Multiplexed Comparative Sequence Analysis) by employing base-specific cleavage biochemistry and MALDI TOF-MS analysis.
Figures
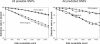



Similar articles
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for genotyping of human platelet-specific antigens.Transfusion. 2009 Feb;49(2):252-8. doi: 10.1111/j.1537-2995.2008.01953.x. Epub 2008 Oct 29. Transfusion. 2009. PMID: 18980617
-
Mass-spectrometry DNA sequencing.Mutat Res. 2005 Jun 3;573(1-2):3-12. doi: 10.1016/j.mrfmmm.2004.07.021. Mutat Res. 2005. PMID: 15829234 Review.
-
Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications.Clin Biochem. 2005 Apr;38(4):335-50. doi: 10.1016/j.clinbiochem.2004.12.005. Clin Biochem. 2005. PMID: 15766735 Review.
-
Typing of single nucleotide polymorphisms by MALDI mass spectrometry: principles and diagnostic applications.Clin Chim Acta. 2006 Jan;363(1-2):95-105. doi: 10.1016/j.cccn.2005.05.040. Epub 2005 Aug 31. Clin Chim Acta. 2006. PMID: 16139255 Review.
-
Rapid genotyping by MALDI-monitored nuclease selection from probe libraries.Nat Biotechnol. 2000 Nov;18(11):1213-6. doi: 10.1038/81226. Nat Biotechnol. 2000. PMID: 11062445
Cited by
-
Novel mutations in VANGL1 in neural tube defects.Hum Mutat. 2009 Jul;30(7):E706-15. doi: 10.1002/humu.21026. Hum Mutat. 2009. PMID: 19319979 Free PMC article.
-
Contribution of VANGL2 mutations to isolated neural tube defects.Clin Genet. 2011 Jul;80(1):76-82. doi: 10.1111/j.1399-0004.2010.01515.x. Epub 2010 Jul 22. Clin Genet. 2011. PMID: 20738329 Free PMC article.
-
Epigenetically induced ectopic expression of UNCX impairs the proliferation and differentiation of myeloid cells.Haematologica. 2017 Jul;102(7):1204-1214. doi: 10.3324/haematol.2016.163022. Epub 2017 Apr 14. Haematologica. 2017. PMID: 28411256 Free PMC article.
-
Genetic analysis of disheveled 2 and disheveled 3 in human neural tube defects.J Mol Neurosci. 2013 Mar;49(3):582-8. doi: 10.1007/s12031-012-9871-9. Epub 2012 Aug 15. J Mol Neurosci. 2013. PMID: 22892949 Free PMC article.
-
The orphan receptor GPR88 controls impulsivity and is a risk factor for Attention-Deficit/Hyperactivity Disorder.Mol Psychiatry. 2022 Nov;27(11):4662-4672. doi: 10.1038/s41380-022-01738-w. Epub 2022 Sep 8. Mol Psychiatry. 2022. PMID: 36075963 Free PMC article.
References
-
- Böcker S. SNP and mutation discovery using base-specific cleavage and MALDI-TOF mass spectrometry. Bioinformatics. 2003;19(Suppl. 1):I44–I53. - PubMed
-
- Storm N., Darnhofer-Patel B., van den Boom D., Rodi C.P. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 2003;212:241–262. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

