Translation of the first upstream ORF in the hepatitis B virus pregenomic RNA modulates translation at the core and polymerase initiation codons
- PMID: 15731337
- PMCID: PMC549565
- DOI: 10.1093/nar/gki251
Translation of the first upstream ORF in the hepatitis B virus pregenomic RNA modulates translation at the core and polymerase initiation codons
Abstract
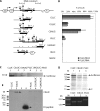
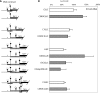
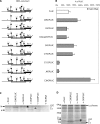
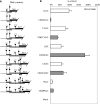
The human hepatitis B virus (HBV) has a compact genome encoding four major overlapping coding regions: the core, polymerase, surface and X. The polymerase initiation codon is preceded by the partially overlapping core and four or more upstream initiation codons. There is evidence that several mechanisms are used to enable the synthesis of the polymerase protein, including leaky scanning and ribosome reinitiation. We have examined the first AUG in the pregenomic RNA, it precedes that of the core. It initiates an uncharacterized short upstream open reading frame (uORF), highly conserved in all HBV subtypes, we designated the C0 ORF. This arrangement suggested that expression of the core and polymerase may be affected by this uORF. Initiation at the C0 ORF was confirmed in reporter constructs in transfected cells. The C0 ORF had an inhibitory role in downstream expression from the core initiation site in HepG2 cells and in vitro, but also stimulated reinitiation at the polymerase start when in an optimal context. Our results indicate that the C0 ORF is a determinant in balancing the synthesis of the core and polymerase proteins.
Figures


Similar articles
-
Expression studies of the HCV-1a core+1 open reading frame in mammalian cells.Virus Res. 2008 May;133(2):123-35. doi: 10.1016/j.virusres.2007.10.019. Epub 2008 Feb 19. Virus Res. 2008. PMID: 18243391
-
Regulation of Tomato golden mosaic virus AL2 and AL3 gene expression by a conserved upstream open reading frame.Virology. 2009 Jan 20;383(2):310-8. doi: 10.1016/j.virol.2008.10.020. Epub 2008 Nov 14. Virology. 2009. PMID: 19012941
-
A sequence required for -1 ribosomal frameshifting located four kilobases downstream of the frameshift site.J Mol Biol. 2001 Jul 27;310(5):987-99. doi: 10.1006/jmbi.2001.4801. J Mol Biol. 2001. PMID: 11502008
-
Translational termination-reinitiation in RNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1558-64. doi: 10.1042/BST0381558. Biochem Soc Trans. 2010. PMID: 21118126 Review.
-
Hepatitis B viruses: reverse transcription a different way.Virus Res. 2008 Jun;134(1-2):235-49. doi: 10.1016/j.virusres.2007.12.024. Epub 2008 Mar 12. Virus Res. 2008. PMID: 18339439 Review.
Cited by
-
An upstream open reading frame modulates ebola virus polymerase translation and virus replication.PLoS Pathog. 2013 Jan;9(1):e1003147. doi: 10.1371/journal.ppat.1003147. Epub 2013 Jan 31. PLoS Pathog. 2013. PMID: 23382680 Free PMC article.
-
Unconventional viral gene expression mechanisms as therapeutic targets.Nature. 2021 May;593(7859):362-371. doi: 10.1038/s41586-021-03511-5. Epub 2021 May 19. Nature. 2021. PMID: 34012080 Review.
-
Characterization of the pleiotropic effects of the genotype G-specific 36-nucleotide insertion in the context of other hepatitis B virus genotypes.J Virol. 2011 Dec;85(24):13278-89. doi: 10.1128/JVI.05583-11. Epub 2011 Oct 12. J Virol. 2011. PMID: 21994450 Free PMC article.
-
Ribosome Shunting, Polycistronic Translation, and Evasion of Antiviral Defenses in Plant Pararetroviruses and Beyond.Front Microbiol. 2018 Apr 10;9:644. doi: 10.3389/fmicb.2018.00644. eCollection 2018. Front Microbiol. 2018. PMID: 29692761 Free PMC article. Review.
-
Transterm: a database to aid the analysis of regulatory sequences in mRNAs.Nucleic Acids Res. 2009 Jan;37(Database issue):D72-6. doi: 10.1093/nar/gkn763. Epub 2008 Nov 4. Nucleic Acids Res. 2009. PMID: 18984623 Free PMC article.
References
-
- Chin R., Locarnini S. Treatment of chronic hepatitis B: current challenges and future directions. Rev. Med. Virol. 2003;13:255–272. - PubMed
-
- Feld J., Lee J.Y., Locarnini S. New targets and possible new therapeutic approaches in the chemotherapy of chronic hepatitis B. Hepatology. 2003;38:545–553. - PubMed
-
- Ganem D., Varmus H.E. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 1987;56:651–693. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

