The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides
- PMID: 15731338
- PMCID: PMC549566
- DOI: 10.1093/nar/gki257
The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides
Abstract
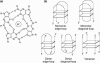
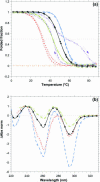
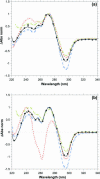
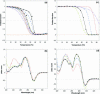
A systematic study of the thermal and conformational properties of chemically modified G-quadruplexes of different molecularities is reported. The effect of backbone charge and atom size, thymine/uracyl substitution as well as the effect of modification at the ribose 2'-position was analyzed by UV spectroscopy. Additional calorimetric studies were performed on different modified forms of the human telomeric sequence. Determination of the differential spectra allowed more insights into the conformational properties of the oligonucleotides. Lack of negative charge at the phosphate backbone yielded to a general destabilization of the G-quadruplex structure. On the other hand, substitution of thymine with uracyl resulted in a moderate or strong stabilization of the structure. Additional modification at the sugar 2'-position gave rise to different effects depending on the molecularity of the quadruplex. In particular, loss of hydrogen bond capacity at the 2'-position strongly affected the conformation of the G-quadruplex. Altogether, these results demonstrate that the effect of some modifications depends on the sequence context, thus providing helpful information for the use of chemically modified quadruplexes as therapeutic agents or as structural elements of supramolecular complexes.
Figures

References
-
- Keniry M. Quadruplex structures in nucleic acids. Biopolymers. 2001;56:123–146. - PubMed
-
- Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. - PubMed
-
- Blackburn E.H. Telomeres: no end in sight. Cell. 1994;77:621–623. - PubMed
-
- Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

