Separation of mutation avoidance and antirecombination functions in an Escherichia coli mutS mutant
- PMID: 15731339
- PMCID: PMC549567
- DOI: 10.1093/nar/gki263
Separation of mutation avoidance and antirecombination functions in an Escherichia coli mutS mutant
Abstract
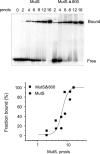
DNA mismatch repair in Escherichia coli has been shown to be involved in two distinct processes: mutation avoidance, which removes potential mutations arising as replication errors, and antirecombination which prevents recombination between related, but not identical (homeologous), DNA sequences. We show that cells with the mutSDelta800 mutation (which removes the C-terminal 53 amino acids of MutS) on a multicopy plasmid are proficient for mutation avoidance. In interspecies genetic crosses, however, recipients with the mutSDelta800 mutation show increased recombination by up to 280-fold relative to mutS+. The MutSDelta800 protein binds to O6-methylguanine mismatches but not to intrastrand platinated GG cross-links, explaining why dam bacteria with the mutSDelta800 mutation are resistant to cisplatin, but not MNNG, toxicity. The results indicate that the C-terminal end of MutS is necessary for antirecombination and cisplatin sensitization, but less significant for mutation avoidance. The inability of MutSDelta800 to form tetramers may indicate that these are the active form of MutS.
Figures


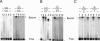

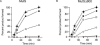
References
-
- Modrich P., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. - PubMed
-
- Schofield M.J., Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. - PubMed
-
- Marti T.M., Kunz C., Fleck O. DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol. 2002;191:28–41. - PubMed
-
- Rayssiguier C., Thaler D.S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

