The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots
- PMID: 15734909
- PMCID: PMC1065410
- DOI: 10.1104/pp.104.057216
The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots
Abstract
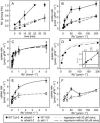
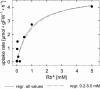
Potassium is an important macronutrient and the most abundant cation in plants. Because soil mineral conditions can vary, plants must be able to adjust to different nutrient availabilities. Here, we used Affymetrix Genechip microarrays to identify genes responsive to potassium (K(+)) deprivation in roots of mature Arabidopsis (Arabidopsis thaliana) plants. Unexpectedly, only a few genes were changed in their expression level after 6, 48, and 96 h of K(+) starvation even though root K(+) content was reduced by approximately 60%. AtHAK5, a potassium transporter gene from the KUP/HAK/KT family, was most consistently and strongly up-regulated in its expression level across 48-h, 96-h, and 7-d K(+) deprivation experiments. AtHAK5 promoter-beta-glucuronidase and -green fluorescent protein fusions showed AtHAK5 promoter activity in the epidermis and vasculature of K(+) deprived roots. Rb(+) uptake kinetics in roots of athak5 T-DNA insertion mutants and wild-type plants demonstrated the absence of a major part of an inducible high-affinity Rb(+)/K(+) (K(m) approximately 15-24 microm) transport system in athak5 plants. In comparative analyses, uptake kinetics of the K(+) channel mutant akt1-1 showed that akt1-1 roots are mainly impaired in a major transport mechanism, with an apparent affinity of approximately 0.9 mm K(+)(Rb(+)). Data show adaptation of apparent K(+) affinities of Arabidopsis roots when individual K(+) transporter genes are disrupted. In addition, the limited transcriptome-wide response to K(+) starvation indicates that posttranscriptional mechanisms may play important roles in root adaptation to K(+) availability in Arabidopsis. The results demonstrate an in vivo function for AtHAK5 in the inducible high-affinity K(+) uptake system in Arabidopsis roots.
Figures

References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

