Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis
- PMID: 15734910
- PMCID: PMC1065389
- DOI: 10.1104/pp.104.056747
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis
Abstract
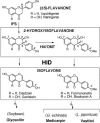
Isoflavonoids are ecophysiologically active secondary metabolites of the Leguminosae and known for health-promoting phytoestrogenic functions. Isoflavones are synthesized by 1,2-elimination of water from 2-hydroxyisoflavanones, the first intermediate with the isoflavonoid skeleton, but details of this dehydration have been unclear. We screened the extracts of repeatedly fractionated Escherichia coli expressing a Glycyrrhiza echinata cDNA library for the activity to convert a radiolabeled precursor into formononetin (7-hydroxy-4'-methoxyisoflavone), and a clone of 2-hydroxyisoflavanone dehydratase (HID) was isolated. Another HID cDNA was cloned from soybean (Glycine max), based on the sequence information in its expressed sequence tag library. Kinetic studies revealed that G. echinata HID is specific to 2,7-dihydroxy-4'-methoxyisoflavanone, while soybean HID has broader specificity to both 4'-hydroxylated and 4'-methoxylated 2-hydroxyisoflavanones, reflecting the structures of isoflavones contained in each plant species. Strikingly, HID proteins were members of a large carboxylesterase family, of which plant proteins form a monophyletic group and some are assigned defensive functions with no intrinsic catalytic activities identified. Site-directed mutagenesis with soybean HID protein suggested that the characteristic oxyanion hole and catalytic triad are essential for the dehydratase as well as the faint esterase activities. The findings, to our knowledge, represent a new example of recruitment of enzymes of primary metabolism during the molecular evolution of plant secondary metabolism.
Figures

References
-
- Akashi T, Saito N, Hirota H, Ayabe S (1997) Anthocyanin-producing dandelion callus as a chalcone synthase source in recombinant polyketide reductase assay. Phytochemistry 46: 283–287 - PubMed
-
- Akashi T, Sawada Y, Aoki T, Ayabe S (2000) New scheme of the biosynthesis of formononetin involving 2,7,4′-trihydroxyisoflavanone but not daidzein as the methyl acceptor. Biosci Biotechnol Biochem 64: 2276–2279 - PubMed
-
- Akashi T, Sawada Y, Shimada N, Sakurai N, Aoki T, Ayabe S (2003) cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol 44: 103–112 - PubMed
-
- Aoki T, Akashi T, Ayabe S (2000) Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J Plant Res 113: 475–488
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

