Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals
- PMID: 15735639
- PMCID: PMC2923337
- DOI: 10.1038/nature03441
Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals
Abstract
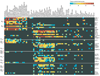
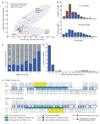
Comprehensive identification of all functional elements encoded in the human genome is a fundamental need in biomedical research. Here, we present a comparative analysis of the human, mouse, rat and dog genomes to create a systematic catalogue of common regulatory motifs in promoters and 3' untranslated regions (3' UTRs). The promoter analysis yields 174 candidate motifs, including most previously known transcription-factor binding sites and 105 new motifs. The 3'-UTR analysis yields 106 motifs likely to be involved in post-transcriptional regulation. Nearly one-half are associated with microRNAs (miRNAs), leading to the discovery of many new miRNA genes and their likely target genes. Our results suggest that previous estimates of the number of human miRNA genes were low, and that miRNAs regulate at least 20% of human genes. The overall results provide a systematic view of gene regulation in the human, which will be refined as additional mammalian genomes become available.
Figures


References
-
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. - PubMed
-
- Pennacchio LA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. - PubMed
-
- Boffelli D, et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

