Engineering gene networks to emulate Drosophila embryonic pattern formation
- PMID: 15736977
- PMCID: PMC1044831
- DOI: 10.1371/journal.pbio.0030064
Engineering gene networks to emulate Drosophila embryonic pattern formation
Abstract
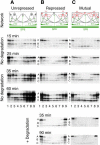
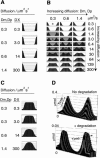
Pattern formation is essential in the development of higher eukaryotes. For example, in the Drosophila embryo, maternal morphogen gradients establish gap gene expression domain patterning along the anterior-posterior axis, through linkage with an elaborate gene network. To understand the evolution and behaviour of such systems better, it is important to establish the minimal determinants required for patterning. We have therefore engineered artificial transcription-translation networks that generate simple patterns, crudely analogous to the Drosophila gap gene system. The Drosophila syncytium was modelled using DNA-coated paramagnetic beads fixed by magnets in an artificial chamber, forming a gene expression network. Transient expression domain patterns were generated using various levels of network connectivity. Generally, adding more transcription repression interactions increased the "sharpness" of the pattern while reducing overall expression levels. An accompanying computer model for our system allowed us to search for parameter sets compatible with patterning. While it is clear that the Drosophila embryo is far more complex than our simplified model, several features of interest emerge. For example, the model suggests that simple diffusion may be too rapid for Drosophila-scale patterning, implying that sublocalisation, or "trapping," is required. Second, we find that for pattern formation to occur under the conditions of our in vitro reaction-diffusion system, the activator molecules must propagate faster than the inhibitors. Third, adding controlled protease degradation to the system stabilizes pattern formation over time. We have reconstituted transcriptional pattern formation from purified substances, including phage RNA polymerases, ribonucleotides, and an eukaryotic translation extract. We anticipate that the system described here will be generally applicable to the study of any biological network with a spatial component.
Figures



References
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. - PubMed
-
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli . Nature. 2000;403:339–342. - PubMed
-
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. - PubMed
-
- Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. - PubMed
-
- Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

