Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis
- PMID: 15738054
- PMCID: PMC2212830
- DOI: 10.1084/jem.20042034
Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis
Abstract
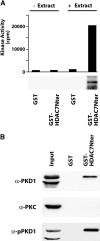
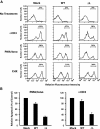
The molecular basis of thymocyte negative selection, a crucial mechanism in establishing central tolerance, is not yet resolved. Histone deacetylases (HDACs) have emerged as key transcriptional regulators in several major developmental programs. Recently, we showed that the class IIa member, HDAC7, regulates negative selection by repressing expression of Nur77, an orphan nuclear receptor involved in antigen-induced apoptosis of thymocytes. Engagement of the T cell receptor (TCR) alleviates this repression through phosphorylation-dependent nuclear exclusion of HDAC7. However, the identity of the TCR-activated kinase that phosphorylates and inactivates HDAC7 was still unknown. Here, we demonstrate that TCR-induced nuclear export of HDAC7 and Nur77 expression is mediated by activation of protein kinase D (PKD). Indeed, active PKD stimulates HDAC7 nuclear export and Nur77 expression. In contrast, inhibition of PKD prevents TCR-mediated nuclear exclusion of HDAC7 and associated Nur77 activation. Furthermore, we show that HDAC7 is an interaction partner and a substrate for PKD. We identify four serine residues in the NH(2) terminus of HDAC7 as targets for PKD. More importantly, a mutant of HDAC7 specifically deficient in phosphorylation by PKD, inhibits TCR-mediated apoptosis of T cell hybridomas. These findings indicate that PKD is likely to play a key role in the signaling pathways controlling negative selection.
Figures






References
-
- Sprent, J., and H. Kishimoto. 2002. The thymus and negative selection. Immunol. Rev. 185: 126–135. - PubMed
-
- Hogquist, K.A. 2001. Signal strength in thymic selection and lineage commitment. Curr. Opin. Immunol. 13:225–231. - PubMed
-
- Starr, T.K., S.C. Jameson, and K.A. Hogquist. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176. - PubMed
-
- Berg, L.J., and J. Kang. 2001. Molecular determinants of TCR expression and selection. Curr. Opin. Immunol. 13:232–241. - PubMed
-
- Winoto, A. 1997. Genes involved in T-cell receptor-mediated apoptosis of thymocytes and T-cell hybridomas. Semin. Immunol. 9:51–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

