Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock
- PMID: 15738418
- PMCID: PMC553333
- DOI: 10.1073/pnas.0409889102
Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock
Abstract
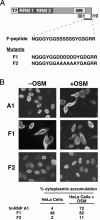
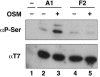
Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 is an alternative splicing factor that is mainly nuclear, although it shuttles rapidly between nuclear and cytoplasmic compartments. Cells stressed by osmotic shock (OSM) activate the mitogen-activated protein kinase kinase(3/6)-p38 signaling pathway, which in turn results in accumulation of hnRNP A1 in the cytoplasm. This effect modulates alternative splicing regulation in vivo and correlates with increased hnRNP A1 phosphorylation. We have characterized the molecular mechanism involved in the cytoplasmic accumulation of hnRNP A1 in NIH 3T3 cells subjected to OSM. This treatment results in serine-specific phosphorylation within a C-terminal peptide, dubbed the "F-peptide," which is adjacent to the M9 motif that mediates bidirectional transport of hnRNP A1. Analysis of mutants in which the F-peptide serines were replaced by aspartic acids or alanines showed that F-peptide phosphorylation is required for the subcellular redistribution of hnRNP A1 in cells subjected to OSM. Furthermore, F-peptide phosphorylation modulates the interaction of hnRNP A1 with transportin Trn1. Our findings suggest that the phosphorylation of F-peptide by cell-signaling pathways regulates the rate of hnRNP A1 nuclear import.
Figures


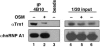

Similar articles
-
The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation.J Cell Biol. 2000 Apr 17;149(2):307-16. doi: 10.1083/jcb.149.2.307. J Cell Biol. 2000. PMID: 10769024 Free PMC article.
-
Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins.J Cell Biol. 1997 Sep 22;138(6):1181-92. doi: 10.1083/jcb.138.6.1181. J Cell Biol. 1997. PMID: 9298975 Free PMC article.
-
Two motifs essential for nuclear import of the hnRNP A1 nucleocytoplasmic shuttling sequence M9 core.FEBS Lett. 2006 Feb 20;580(5):1365-70. doi: 10.1016/j.febslet.2006.01.058. Epub 2006 Jan 26. FEBS Lett. 2006. PMID: 16455081
-
Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles.Mol Cell Neurosci. 2013 Sep;56:436-46. doi: 10.1016/j.mcn.2012.12.002. Epub 2012 Dec 14. Mol Cell Neurosci. 2013. PMID: 23247072 Review.
-
hnRNP A1: the Swiss army knife of gene expression.Int J Mol Sci. 2013 Sep 16;14(9):18999-9024. doi: 10.3390/ijms140918999. Int J Mol Sci. 2013. PMID: 24065100 Free PMC article. Review.
Cited by
-
A Comprehensive Analysis of the Role of hnRNP A1 Function and Dysfunction in the Pathogenesis of Neurodegenerative Disease.Front Mol Biosci. 2021 Apr 12;8:659610. doi: 10.3389/fmolb.2021.659610. eCollection 2021. Front Mol Biosci. 2021. PMID: 33912591 Free PMC article. Review.
-
Alternative Splicing of the Pituitary Adenylate Cyclase-Activating Polypeptide Receptor PAC1: Mechanisms of Fine Tuning of Brain Activity.Front Endocrinol (Lausanne). 2013 May 21;4:55. doi: 10.3389/fendo.2013.00055. eCollection 2013. Front Endocrinol (Lausanne). 2013. PMID: 23734144 Free PMC article.
-
Cell stress modulates the function of splicing regulatory protein RBM4 in translation control.Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2235-40. doi: 10.1073/pnas.0611015104. Epub 2007 Feb 6. Proc Natl Acad Sci U S A. 2007. PMID: 17284590 Free PMC article.
-
Natural and pathogenic protein sequence variation affecting prion-like domains within and across human proteomes.BMC Genomics. 2020 Jan 8;21(1):23. doi: 10.1186/s12864-019-6425-3. BMC Genomics. 2020. PMID: 31914925 Free PMC article.
-
The Splicing Factor hnRNP M Is a Critical Regulator of Innate Immune Gene Expression in Macrophages.Cell Rep. 2019 Nov 5;29(6):1594-1609.e5. doi: 10.1016/j.celrep.2019.09.078. Cell Rep. 2019. PMID: 31693898 Free PMC article.
References
-
- Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195-205. - PubMed
-
- Krecic, A. M. & Swanson, M. S. (1999) Curr. Opin. Cell Biol. 11, 363-371. - PubMed
-
- Piñol-Roma, S., Choi, Y. D., Matunis, M. J. & Dreyfuss, G. (1988) Genes Dev. 2, 215-227. - PubMed
-
- Dreyfuss, G., Matunis, M. J., Piñol-Roma, S. & Burd, C. G. (1993) Annu. Rev. Biochem. 62, 289-321. - PubMed
-
- Michael, W. M., Siomi, H., Choi, M., Piñol-Roma, S., Nakielny, S., Liu, Q. & Dreyfuss, G. (1995) Cold Spring Harb. Symp. Quant. Biol. 60, 663-668. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

