The contribution of phosphate-phosphate repulsions to the free energy of DNA bending
- PMID: 15741179
- PMCID: PMC552960
- DOI: 10.1093/nar/gki272
The contribution of phosphate-phosphate repulsions to the free energy of DNA bending
Abstract
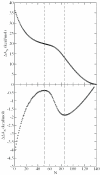
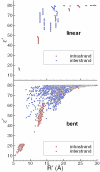
DNA bending is important for the packaging of genetic material, regulation of gene expression and interaction of nucleic acids with proteins. Consequently, it is of considerable interest to quantify the energetic factors that must be overcome to induce bending of DNA, such as base stacking and phosphate-phosphate repulsions. In the present work, the electrostatic contribution of phosphate-phosphate repulsions to the free energy of bending DNA is examined for 71 bp linear and bent-form model structures. The bent DNA model was based on the crystallographic structure of a full turn of DNA in a nucleosome core particle. A Green's function approach based on a linear-scaling smooth conductor-like screening model was applied to ascertain the contribution of individual phosphate-phosphate repulsions and overall electrostatic stabilization in aqueous solution. The effect of charge neutralization by site-bound ions was considered using Monte Carlo simulation to characterize the distribution of ion occupations and contribution of phosphate repulsions to the free energy of bending as a function of counterion load. The calculations predict that the phosphate-phosphate repulsions account for approximately 30% of the total free energy required to bend DNA from canonical linear B-form into the conformation found in the nucleosome core particle.
Figures


References
-
- Bloomfield V.A., Crothers D.M., Tinoco I., Jr . Nucleic Acids: Structures, Properties, and Functions. Sausalito, CA: University Science Books; 2000.
-
- Olson W.K., Zhurkin V.B. Modeling DNA deformations. Curr. Opin. Struct. Biol. 2000;10:286–297. - PubMed
-
- Horton N.C., Perona J.J. DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites. Biochemistry. 2004;43:6841–6857. - PubMed

