Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control
- PMID: 15741314
- PMCID: PMC1065720
- DOI: 10.1101/gad.1279805
Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control
Abstract
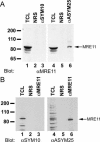
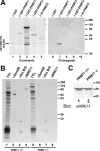
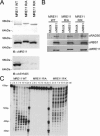
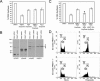
The role of protein arginine methylation in the DNA damage checkpoint response and DNA repair is largely unknown. Herein we show that the MRE11 checkpoint protein is arginine methylated by PRMT1. Mutation of the arginines within MRE11 severely impaired the exonuclease activity of MRE11 but did not influence its ability to form complexes with RAD50 and NBS1. Cells containing hypomethylated MRE11 displayed intra-S-phase DNA damage checkpoint defects that were significantly rescued with the MRE11-RAD50-NBS1 complex. Our results suggest that arginine methylation regulates the activity of MRE11-RAD50-NBS1 complex during the intra-S-phase DNA damage checkpoint response.
Figures
References
-
- Bedford M.T., Frankel, A., Yaffe, M.B., Clarke, S., Leder, P., and Richard, S. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275: 16030-16036. - PubMed
-
- Boisvert F.M., Cote, J., Boulanger, M.C., and Richard, S. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2: 1319-1330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
