Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR
- PMID: 15741343
- PMCID: PMC2253452
- DOI: 10.1110/ps.041231005
Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR
Abstract
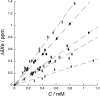
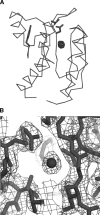
The chemical shift of the (129)Xe NMR signal has been shown to be extremely sensitive to the local environment around the atom and has been used to follow processes such as ligand binding by bacterial periplasmic binding proteins. Here we show that the (129)Xe shift can sense more subtle changes: magnesium binding, BeF(3)(-) activation, and peptide binding by the Escherichia coli chemotaxis Y protein. (1)H-(15)N correlation spectroscopy and X-ray crystallography were used to identify two xenon-binding cavities in CheY that are primarily responsible for the shift changes. One site is near the active site, and the other is near the peptide binding site.
Figures


Comment in
-
Detection of multiple protein conformations by laser-polarized xenon.Protein Sci. 2005 Apr;14(4):847. doi: 10.1110/ps.051398705. Protein Sci. 2005. PMID: 15772304 Free PMC article. No abstract available.
Similar articles
-
Detection of multiple protein conformations by laser-polarized xenon.Protein Sci. 2005 Apr;14(4):847. doi: 10.1110/ps.051398705. Protein Sci. 2005. PMID: 15772304 Free PMC article. No abstract available.
-
Structural investigation of pig metmyoglobin by 129Xe NMR spectroscopy.Biochim Biophys Acta. 2004 Sep 24;1674(2):182-92. doi: 10.1016/j.bbagen.2004.06.011. Biochim Biophys Acta. 2004. PMID: 15374622
-
Nuclear magnetic resonance spectroscopy reveals the functional state of the signalling protein CheY in vivo in Escherichia coli.Mol Microbiol. 2003 Sep;49(5):1191-200. doi: 10.1046/j.1365-2958.2003.03628.x. Mol Microbiol. 2003. PMID: 12940980
-
Use of 19F NMR to probe protein structure and conformational changes.Annu Rev Biophys Biomol Struct. 1996;25:163-95. doi: 10.1146/annurev.bb.25.060196.001115. Annu Rev Biophys Biomol Struct. 1996. PMID: 8800468 Free PMC article. Review.
-
Response regulation in bacterial chemotaxis.J Cell Biochem. 1993 Jan;51(1):41-6. doi: 10.1002/jcb.240510109. J Cell Biochem. 1993. PMID: 8381790 Review.
Cited by
-
Insights into correlated motions and long-range interactions in CheY derived from molecular dynamics simulations.Biophys J. 2007 Mar 15;92(6):2062-79. doi: 10.1529/biophysj.106.081950. Epub 2006 Dec 15. Biophys J. 2007. PMID: 17172298 Free PMC article.
-
Xenon-Protein Interactions: Characterization by X-Ray Crystallography and Hyper-CEST NMR.Methods Enzymol. 2018;602:249-272. doi: 10.1016/bs.mie.2018.02.005. Epub 2018 Mar 15. Methods Enzymol. 2018. PMID: 29588032 Free PMC article.
-
NMR Hyperpolarization Techniques of Gases.Chemistry. 2017 Jan 18;23(4):725-751. doi: 10.1002/chem.201603884. Epub 2016 Dec 5. Chemistry. 2017. PMID: 27711999 Free PMC article. Review.
-
Hyperpolarized krypton-83 as a contrast agent for magnetic resonance imaging.Proc Natl Acad Sci U S A. 2005 Dec 20;102(51):18275-9. doi: 10.1073/pnas.0509419102. Epub 2005 Dec 12. Proc Natl Acad Sci U S A. 2005. PMID: 16344474 Free PMC article.
-
Capturing cooperative interactions with the PSI-MI format.Database (Oxford). 2013 Sep 25;2013:bat066. doi: 10.1093/database/bat066. Print 2013. Database (Oxford). 2013. PMID: 24067240 Free PMC article.
References
-
- Bellsolell, L., Prieto, J., Serrano, L., and Coll, M. 1994. Mg2+ binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J. Mol. Biol. 238 489–495. - PubMed
-
- Bolotovsky, R., Steller, I., and Rossman, M.G. 1998. The use of partial reflections for scaling and averaging X-ray area-detector data. J. Appl. Crystallogr. 31 708–717.
-
- Bren, A. and Eisenbach, M. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 278 507–514. - PubMed
-
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macro-molecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. - PubMed

