Developmental changes in abundance of the VSPbeta protein following nuclear transformation of maize with the soybean vspbeta cDNA
- PMID: 15743517
- PMCID: PMC554770
- DOI: 10.1186/1471-2229-5-3
Developmental changes in abundance of the VSPbeta protein following nuclear transformation of maize with the soybean vspbeta cDNA
Abstract
Background: Developing monocots that accumulate more vegetative tissue protein is one strategy for improving nitrogen-sequestration and nutritive value of forage and silage crops. In soybeans (a dicotyledonous legume), the vspA and B genes encode subunits of a dimeric vegetative storage protein that plays an important role in nitrogen storage in vegetative tissues. Similar genes are found in monocots; however, they do not accumulate in leaves as storage proteins, and the ability of monocot leaves to support accumulation of an ectopically expressed soybean VSP is in question. To test this, transgenic maize (Zea Mays L. Hi-II hybrid) lines were created expressing soybean vspB from a maize ubiquitin Ubi-1 promoter.
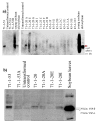
Results: From 81 bombardments, 101 plants were regenerated, and plants from five independent lines produced vspB transcripts and VSPbeta polypeptides. In leaves from seven-week-old plants (prior to flowering), VSPbeta accumulated to 0.5% of the soluble leaf protein in primary transgenic plants (R0), but to only 0.03% in R1 plants. During seed-filling (silage-stage) in R1 plants, the VSPbeta protein was no longer detected in leaves and stems despite continued presence of the vspB RNA. The RNA transcripts for this peptide either became less efficiently translated, or the VSPbeta protein became unstable during seed-fill.
Conclusion: Developmental differences in the accumulation of soybean VSPbeta when transgenically expressed in maize show that despite no changes in the vspB transcript level, VSPbeta protein that is readily detected in leaves of preflowering plants, becomes undetectable as seeds begin to develop.
Figures

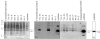
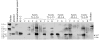

Similar articles
-
Coexpression of the soybean vegetative storage protein beta subunit (S-VSPbeta) either with the bacterial feedback-insensitive dihydrodipicolinate synthase or with S-VSPalpha stabilizes the S-VSPbeta transgene protein and enhances lysine production in transgenic tobacco plants.Transgenic Res. 2003 Feb;12(1):123-6. doi: 10.1023/a:1022130100493. Transgenic Res. 2003. PMID: 12650531
-
[Expression activity of maize Ubi-1 promoter in fertile transgenic maize plants].Shi Yan Sheng Wu Xue Bao. 2002 Dec;35(4):296-302. Shi Yan Sheng Wu Xue Bao. 2002. PMID: 15346987 Chinese.
-
[Transgenic maize plants with low copy number of foreign genes were produced with maize Ubi-1 promoter].Sheng Wu Gong Cheng Xue Bao. 2004 Jan;20(1):120-5. Sheng Wu Gong Cheng Xue Bao. 2004. PMID: 16108502 Chinese.
-
Seed-specific expression of the lysine-rich protein gene sb401 significantly increases both lysine and total protein content in maize seeds.Food Nutr Bull. 2005 Dec;26(4):427-31. Food Nutr Bull. 2005. PMID: 16465991 Review.
-
Plant morphogenesis. More knots untied.Curr Biol. 1994 Jun 1;4(6):529-31. doi: 10.1016/s0960-9822(00)00115-9. Curr Biol. 1994. PMID: 7922374 Review.
Cited by
-
A vegetative storage protein improves drought tolerance in maize.Plant Biotechnol J. 2022 Feb;20(2):374-389. doi: 10.1111/pbi.13720. Epub 2021 Dec 8. Plant Biotechnol J. 2022. PMID: 34614273 Free PMC article.
References
-
- Vogel KP, Sleper DA. Alteration of plant via genetics and plant breeding. In: Faher GC, editor. Forage Quality, Evaluation, and Utilization. Madison, WI, USA: American Society of Agronomy; 1994. pp. 891–921.
-
- Higgins TJV, Spencer D. The expression of a chimeric cauliflower mosaic virus (CaMV 35S)-pea vicilin gene in tobacco. Plant Sci. 1991;74:89–98. doi: 10.1016/0168-9452(91)90259-B. - DOI
-
- Wandelt CI, Knibb W, Schroeder HE, Khan MRI, Spencer D, Craig S, Higgins TJV. The expression of an ovalbumin and a seed protein gene in the leaves of transgenic plants. In: Herman RG, Larkins B, editor. Plant Molecular Biology 2. New York: Plenum Press; 1991. pp. 471–478.
-
- Lawton MA, Tierney MA, Nakamura I, Anderson E, Komeda Y, Dube P, Hoffman N, Fraley RT, Beachy RN. Expression of a soybean β-conglycinin gene under the control of the cauliflower mosaic virus 35S and 19S promoters in transformed petunia tissues. Plant Mol Biol. 1987;9:315–324. doi: 10.1007/BF00014906. - DOI - PubMed
-
- Tabe LM, Higgins CM, McNabb WC, Higgins TJV. Genetic engineering of grain and pasture legumes for improved nutritive value. Genetica. 1993;90:181–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

