Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families
- PMID: 15743757
- PMCID: PMC2932481
- DOI: 10.1074/jbc.M502004200
Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families
Abstract
Gonadotropin-releasing hormone (GnRH) is the central regulator of the hypothalamic-pituitary-gonadal axis, controlling sexual maturation and fertility in diverse species from fish to humans. GnRH gene expression is limited to a discrete population of neurons that migrate through the nasal region into the hypothalamus during embryonic development. The GnRH regulatory region contains four conserved homeodomain binding sites (ATTA) that are essential for basal promoter activity and cell-specific expression of the GnRH gene. MSX and DLX are members of the Antennapedia class of non-Hox homeodomain transcription factors that regulate gene expression and influence development of the craniofacial structures and anterior forebrain. Here, we report that expression patterns of the Msx and Dlx families of homeodomain transcription factors largely coincide with the migratory route of GnRH neurons and co-express with GnRH in neurons during embryonic development. In addition, MSX and DLX family members bind directly to the ATTA consensus sequences and regulate transcriptional activity of the GnRH promoter. Finally, mice lacking MSX1 or DLX1 and 2 show altered numbers of GnRH-expressing cells in regions where these factors likely function. These findings strongly support a role for MSX and DLX in contributing to spatiotemporal regulation of GnRH transcription during development.
Figures

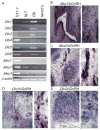

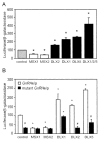
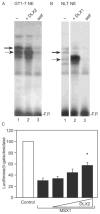


Similar articles
-
TALE homeodomain proteins regulate gonadotropin-releasing hormone gene expression independently and via interactions with Oct-1.J Biol Chem. 2004 Jul 16;279(29):30287-97. doi: 10.1074/jbc.M402960200. Epub 2004 May 11. J Biol Chem. 2004. PMID: 15138251 Free PMC article.
-
Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6.J Neurosci. 2011 Jan 12;31(2):426-38. doi: 10.1523/JNEUROSCI.1688-10.2011. J Neurosci. 2011. PMID: 21228153 Free PMC article.
-
Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors.J Biol Chem. 2009 Jun 19;284(25):16966-16978. doi: 10.1074/jbc.M109.002485. Epub 2009 Apr 28. J Biol Chem. 2009. PMID: 19401468 Free PMC article.
-
Regulation of gonadotropin-releasing hormone gene expression.Semin Reprod Med. 2007 Sep;25(5):313-25. doi: 10.1055/s-2007-984737. Semin Reprod Med. 2007. PMID: 17710727 Review.
-
Gonadotropin-releasing hormone: regulation of the GnRH gene.FEBS J. 2008 Nov;275(22):5458-78. doi: 10.1111/j.1742-4658.2008.06676.x. FEBS J. 2008. PMID: 18959737 Review.
Cited by
-
Homeodomain Proteins SIX3 and SIX6 Regulate Gonadotrope-specific Genes During Pituitary Development.Mol Endocrinol. 2015 Jun;29(6):842-55. doi: 10.1210/me.2014-1279. Epub 2015 Apr 27. Mol Endocrinol. 2015. PMID: 25915183 Free PMC article.
-
Transcriptional regulation of cranial sensory placode development.Curr Top Dev Biol. 2015;111:301-50. doi: 10.1016/bs.ctdb.2014.11.009. Epub 2015 Jan 22. Curr Top Dev Biol. 2015. PMID: 25662264 Free PMC article. Review.
-
Specification of GnRH-1 neurons by antagonistic FGF and retinoic acid signaling.Dev Biol. 2012 Feb 15;362(2):254-62. doi: 10.1016/j.ydbio.2011.12.016. Epub 2011 Dec 19. Dev Biol. 2012. PMID: 22200593 Free PMC article.
-
Patterning, specification, and differentiation in the developing hypothalamus.Wiley Interdiscip Rev Dev Biol. 2015 Sep-Oct;4(5):445-68. doi: 10.1002/wdev.187. Epub 2015 Mar 27. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25820448 Free PMC article. Review.
-
Epigenetic changes coincide with in vitro primate GnRH neuronal maturation.Endocrinology. 2010 Nov;151(11):5359-68. doi: 10.1210/en.2010-0555. Epub 2010 Sep 22. Endocrinology. 2010. PMID: 20861233 Free PMC article.
References
-
- Wu TJ, Gibson MJ, Silverman AJ. J Neuroendocrinol. 1995;7:899–902. - PubMed
-
- Mason AJ, Hayflick JS, Zoeller RT, Young WS, Phillips HS, Nikolics K, Seeburg PH. Science. 1986;234:1366–1371. - PubMed
-
- Dubois EA, Zandbergen MA, Peute J, Goos HJ. Brain Res Bull. 2002;57:413–418. - PubMed
-
- Schwanzel-Fukuda M, Pfaff DW. Nature. 1989;338:161–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

