Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta
- PMID: 15743815
- PMCID: PMC1061627
- DOI: 10.1128/MCB.25.6.2169-2176.2005
Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta
Abstract
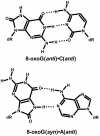
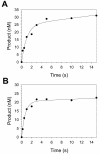
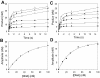
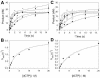
Most DNA polymerases incorporate nucleotides opposite template 7,8-dihydro-8-oxoguanine (8-oxoG) lesions with reduced efficiency and accuracy. DNA polymerase (Pol) eta, which catalyzes the error-free replication of template thymine-thymine (TT) dimers, has the unique ability to accurately and efficiently incorporate nucleotides opposite 8-oxoG templates. Here we have used pre-steady-state kinetics to examine the mechanisms of correct and incorrect nucleotide incorporation opposite G and 8-oxoG by Saccharomyces cerevisiae Pol eta. We found that Pol eta binds the incoming correct dCTP opposite both G and 8-oxoG with similar affinities, and it incorporates the correct nucleotide bound opposite both G and 8-oxoG with similar rates. While Pol eta incorporates an incorrect A opposite 8-oxoG with lower efficiency than it incorporates a correct C, it does incorporate A more efficiently opposite 8-oxoG than opposite G. This is mainly due to greater binding affinity for the incorrect incoming dATP opposite 8-oxoG. Overall, these results show that Pol eta replicates through 8-oxoG without any barriers introduced by the presence of the lesion.
Figures

Similar articles
-
The N2-ethylguanine and the O6-ethyl- and O6-methylguanine lesions in DNA: contrasting responses from the "bypass" DNA polymerase eta and the replicative DNA polymerase alpha.Chem Res Toxicol. 2003 Dec;16(12):1616-23. doi: 10.1021/tx034164f. Chem Res Toxicol. 2003. PMID: 14680376
-
Kinetics of dCTP incorporation opposite to 7,8-dihydro-8-oxoguanine with different 5' nearest neighbors by yeast polymerase eta.Nucleic Acids Symp Ser (Oxf). 2008;(52):531-2. doi: 10.1093/nass/nrn269. Nucleic Acids Symp Ser (Oxf). 2008. PMID: 18776488
-
Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics.Biochemistry. 1997 May 27;36(21):6475-87. doi: 10.1021/bi9627267. Biochemistry. 1997. PMID: 9174365
-
DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions.Curr Opin Struct Biol. 2011 Jun;21(3):358-69. doi: 10.1016/j.sbi.2011.03.008. Epub 2011 Apr 7. Curr Opin Struct Biol. 2011. PMID: 21482102 Free PMC article. Review.
-
A Role for N6-Methyladenine in DNA Damage Repair.Trends Biochem Sci. 2021 Mar;46(3):175-183. doi: 10.1016/j.tibs.2020.09.007. Epub 2020 Oct 16. Trends Biochem Sci. 2021. PMID: 33077363 Free PMC article. Review.
Cited by
-
Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.Structure. 2009 May 13;17(5):725-36. doi: 10.1016/j.str.2009.03.011. Structure. 2009. PMID: 19446528 Free PMC article.
-
Mutagenesis mechanism of the major oxidative adenine lesion 7,8-dihydro-8-oxoadenine.Nucleic Acids Res. 2020 May 21;48(9):5119-5134. doi: 10.1093/nar/gkaa193. Nucleic Acids Res. 2020. PMID: 32282906 Free PMC article.
-
Recent insight into the kinetic mechanisms and conformational dynamics of Y-Family DNA polymerases.Biochemistry. 2014 May 6;53(17):2804-14. doi: 10.1021/bi5000405. Epub 2014 Apr 23. Biochemistry. 2014. PMID: 24716482 Free PMC article. Review.
-
Active Site Interactions Impact Phosphoryl Transfer during Replication of Damaged and Undamaged DNA by Escherichia coli DNA Polymerase I.Chem Res Toxicol. 2017 Nov 20;30(11):2033-2043. doi: 10.1021/acs.chemrestox.7b00257. Epub 2017 Oct 25. Chem Res Toxicol. 2017. PMID: 29053918 Free PMC article.
-
Analyzing the Catalytic Activities and Interactions of Eukaryotic Translesion Synthesis Polymerases.Methods Enzymol. 2017;592:329-356. doi: 10.1016/bs.mie.2017.04.002. Epub 2017 May 8. Methods Enzymol. 2017. PMID: 28668126 Free PMC article.
References
-
- Beckman, K. B., and B. N. Ames. 1997. Oxidative decay of DNA. J. Biol. Chem. 272:19633-19636. - PubMed
-
- Capson, T. L., J. A. Peliska, B. F. Kaboord, M. W. Frey, C. Lively, M. Dahlberg, and S. J. Benkovic. 1992. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry 31:10984-10994. - PubMed
-
- Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262:232-256. - PubMed
-
- Dahlberg, M. E., and S. J. Benkovic. 1991. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry 30:4835-4843. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
