Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta
- PMID: 15743815
- PMCID: PMC1061627
- DOI: 10.1128/MCB.25.6.2169-2176.2005
Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta
Abstract
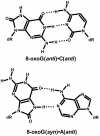
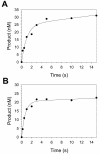
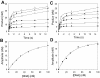
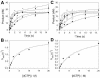
Most DNA polymerases incorporate nucleotides opposite template 7,8-dihydro-8-oxoguanine (8-oxoG) lesions with reduced efficiency and accuracy. DNA polymerase (Pol) eta, which catalyzes the error-free replication of template thymine-thymine (TT) dimers, has the unique ability to accurately and efficiently incorporate nucleotides opposite 8-oxoG templates. Here we have used pre-steady-state kinetics to examine the mechanisms of correct and incorrect nucleotide incorporation opposite G and 8-oxoG by Saccharomyces cerevisiae Pol eta. We found that Pol eta binds the incoming correct dCTP opposite both G and 8-oxoG with similar affinities, and it incorporates the correct nucleotide bound opposite both G and 8-oxoG with similar rates. While Pol eta incorporates an incorrect A opposite 8-oxoG with lower efficiency than it incorporates a correct C, it does incorporate A more efficiently opposite 8-oxoG than opposite G. This is mainly due to greater binding affinity for the incorrect incoming dATP opposite 8-oxoG. Overall, these results show that Pol eta replicates through 8-oxoG without any barriers introduced by the presence of the lesion.
Figures

References
-
- Beckman, K. B., and B. N. Ames. 1997. Oxidative decay of DNA. J. Biol. Chem. 272:19633-19636. - PubMed
-
- Capson, T. L., J. A. Peliska, B. F. Kaboord, M. W. Frey, C. Lively, M. Dahlberg, and S. J. Benkovic. 1992. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry 31:10984-10994. - PubMed
-
- Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262:232-256. - PubMed
-
- Dahlberg, M. E., and S. J. Benkovic. 1991. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry 30:4835-4843. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
