Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20
- PMID: 15743816
- PMCID: PMC1061602
- DOI: 10.1128/MCB.25.6.2177-2190.2005
Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20
Abstract
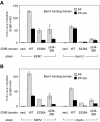
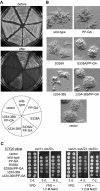
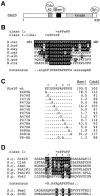
The Saccharomyces cerevisiae PAK (p21-activated kinase) family kinase Ste20 functions in several signal transduction pathways, including pheromone response, filamentous growth, and hyperosmotic resistance. The GTPase Cdc42 localizes and activates Ste20 by binding to an autoinhibitory motif within Ste20 called the CRIB domain. Another factor that functions with Ste20 and Cdc42 is the protein Bem1. Bem1 has two SH3 domains, but target ligands for these domains have not been described. Here we identify an evolutionarily conserved binding site for Bem1 between the CRIB and kinase domains of Ste20. Mutation of tandem proline-rich (PxxP) motifs in this region disrupts Bem1 binding, suggesting that it serves as a ligand for a Bem1 SH3 domain. These PxxP motif mutations affect signaling additively with CRIB domain mutations, indicating that Bem1 and Cdc42 make separable contributions to Ste20 function, which cooperate to promote optimal signaling. This PxxP region also binds another SH3 domain protein, Nbp2, but analysis of bem1Delta versus nbp2Delta strains shows that the signaling defects of PxxP mutants result from impaired binding to Bem1 rather than from impaired binding to Nbp2. Finally, the PxxP mutations also reduce signaling by constitutively active Ste20, suggesting that postactivation functions of PAKs can be promoted by SH3 domain proteins, possibly by colocalizing PAKs with their substrates. The overall results also illustrate how the final signaling function of a protein can be governed by combinatorial addition of multiple, independent protein-protein interaction modules.
Figures




Similar articles
-
Solution structure of a novel Cdc42 binding module of Bem1 and its interaction with Ste20 and Cdc42.J Biol Chem. 2010 Jun 18;285(25):19346-53. doi: 10.1074/jbc.M110.116749. Epub 2010 Apr 21. J Biol Chem. 2010. PMID: 20410294 Free PMC article.
-
A novel Cdc42-interacting domain of the yeast polarity establishment protein Bem1. Implications for modulation of mating pheromone signaling.J Biol Chem. 2007 Jan 5;282(1):29-38. doi: 10.1074/jbc.M609308200. Epub 2006 Nov 7. J Biol Chem. 2007. PMID: 17090539
-
Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20.Mol Cell Biol. 2002 May;22(9):2939-51. doi: 10.1128/MCB.22.9.2939-2951.2002. Mol Cell Biol. 2002. PMID: 11940652 Free PMC article.
-
Pheromone response, mating and cell biology.Curr Opin Microbiol. 2000 Dec;3(6):573-81. doi: 10.1016/s1369-5274(00)00143-0. Curr Opin Microbiol. 2000. PMID: 11121776 Review.
-
Signalosome assembly by domains undergoing dynamic head-to-tail polymerization.Trends Biochem Sci. 2014 Oct;39(10):487-95. doi: 10.1016/j.tibs.2014.08.006. Epub 2014 Sep 16. Trends Biochem Sci. 2014. PMID: 25239056 Review.
Cited by
-
Nucleophosmin is a binding partner of nucleostemin in human osteosarcoma cells.Mol Biol Cell. 2008 Jul;19(7):2870-5. doi: 10.1091/mbc.e08-02-0128. Epub 2008 Apr 30. Mol Biol Cell. 2008. PMID: 18448670 Free PMC article.
-
Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex.Curr Biol. 2008 Nov 25;18(22):1719-26. doi: 10.1016/j.cub.2008.09.060. Epub 2008 Nov 13. Curr Biol. 2008. PMID: 19013066 Free PMC article.
-
A time-resolved interaction analysis of Bem1 reconstructs the flow of Cdc42 during polar growth.Life Sci Alliance. 2020 Jul 31;3(9):e202000813. doi: 10.26508/lsa.202000813. Print 2020 Sep. Life Sci Alliance. 2020. PMID: 32737079 Free PMC article.
-
Solution structure of a novel Cdc42 binding module of Bem1 and its interaction with Ste20 and Cdc42.J Biol Chem. 2010 Jun 18;285(25):19346-53. doi: 10.1074/jbc.M110.116749. Epub 2010 Apr 21. J Biol Chem. 2010. PMID: 20410294 Free PMC article.
-
PathFinder: mining signal transduction pathway segments from protein-protein interaction networks.BMC Bioinformatics. 2007 Sep 13;8:335. doi: 10.1186/1471-2105-8-335. BMC Bioinformatics. 2007. PMID: 17854489 Free PMC article.
References
-
- Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. - PMC - PubMed
-
- Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241-263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
