Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors
- PMID: 15743823
- PMCID: PMC1061617
- DOI: 10.1128/MCB.25.6.2273-2287.2005
Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors
Erratum in
- Mol Cell Biol. 2006 Apr;26(8):3335
Abstract
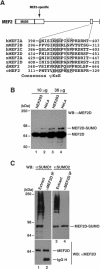
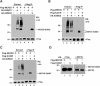
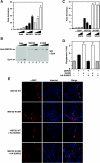
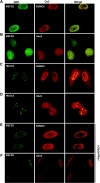
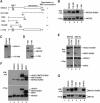
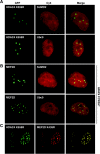
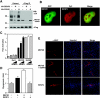
The myocyte enhancer factor-2 (MEF2) family of transcription factors plays an important role in regulating cellular programs like muscle differentiation, neuronal survival, and T-cell apoptosis. Multisite phosphorylation is known to control the transcriptional activity of MEF2 proteins, but it is unclear whether other modifications are involved. Here, we report that human MEF2D, as well as MEF2C, is modified by SUMO2 and SUMO3 at a motif highly conserved among MEF2 proteins from diverse organisms. This motif is located within the C-terminal transcriptional activation domain, and its sumoylation inhibits transcription. As a transcriptional corepressor of MEF2, histone deacetylase 4 (HDAC4) potentiates sumoylation. This potentiation is dependent on the N-terminal region but not the C-terminal deacetylase domain of HDAC4 and is inhibited by the sumoylation of HDAC4 itself. Moreover, HDAC5, HDAC7, and an HDAC9 isoform also stimulate sumoylation of MEF2. Opposing the action of class IIa deacetylases, the SUMO protease SENP3 reverses the sumoylation to augment the transcriptional and myogenic activities of MEF2. Similarly, the calcium/calmodulin-dependent kinases [corrected] and extracellular signal-regulated kinase 5 signaling pathways negatively regulate the sumoylation. These results thus identify sumoylation as a novel regulatory mechanism for MEF2 and suggest that this modification interplays with phosphorylation to promote intramolecular signaling for coordinated regulation in vivo.
Figures
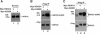


Similar articles
-
Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2.Mol Cell Biol. 2007 Feb;27(4):1280-95. doi: 10.1128/MCB.00882-06. Epub 2006 Dec 11. Mol Cell Biol. 2007. PMID: 17158926 Free PMC article.
-
Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications.Mol Cell Biol. 2005 Oct;25(19):8456-64. doi: 10.1128/MCB.25.19.8456-8464.2005. Mol Cell Biol. 2005. PMID: 16166628 Free PMC article.
-
Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation.J Biol Chem. 2006 Feb 17;281(7):4423-33. doi: 10.1074/jbc.M509471200. Epub 2005 Dec 15. J Biol Chem. 2006. PMID: 16356933
-
Class II histone deacetylases: structure, function, and regulation.Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review.
-
MEF2: a central regulator of diverse developmental programs.Development. 2007 Dec;134(23):4131-40. doi: 10.1242/dev.008367. Epub 2007 Oct 24. Development. 2007. PMID: 17959722 Review.
Cited by
-
The role of SUMOylation during development.Biochem Soc Trans. 2020 Apr 29;48(2):463-478. doi: 10.1042/BST20190390. Biochem Soc Trans. 2020. PMID: 32311032 Free PMC article. Review.
-
The role of histone deacetylase 4 during chondrocyte hypertrophy and endochondral bone development.Bone Joint Res. 2020 May 16;9(2):82-89. doi: 10.1302/2046-3758.92.BJR-2019-0172.R1. eCollection 2020 Feb. Bone Joint Res. 2020. PMID: 32435460 Free PMC article. Review.
-
Interleukin 1β triggers synaptic and memory deficits in Herpes simplex virus type-1-infected mice by downregulating the expression of synaptic plasticity-related genes via the epigenetic MeCP2/HDAC4 complex.Cell Mol Life Sci. 2023 Jun 1;80(6):172. doi: 10.1007/s00018-023-04817-5. Cell Mol Life Sci. 2023. PMID: 37261502 Free PMC article.
-
PIAS1 modulates striatal transcription, DNA damage repair, and SUMOylation with relevance to Huntington's disease.Proc Natl Acad Sci U S A. 2021 Jan 26;118(4):e2021836118. doi: 10.1073/pnas.2021836118. Proc Natl Acad Sci U S A. 2021. PMID: 33468657 Free PMC article.
-
B cells infected with Type 2 Epstein-Barr virus (EBV) have increased NFATc1/NFATc2 activity and enhanced lytic gene expression in comparison to Type 1 EBV infection.PLoS Pathog. 2020 Feb 14;16(2):e1008365. doi: 10.1371/journal.ppat.1008365. eCollection 2020 Feb. PLoS Pathog. 2020. PMID: 32059024 Free PMC article.
References
-
- Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. - PubMed
-
- Bakin, R. E., and M. O. Jung. 2004. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 279:51218-51225. - PubMed
-
- Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. - PubMed
-
- Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
