Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast
- PMID: 15743909
- PMCID: PMC1087237
- DOI: 10.1091/mbc.e04-09-0781
Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast
Abstract
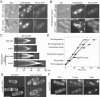
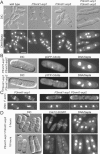
Fission yeast capping protein SpCP is a heterodimer of two subunits (Acp1p and Acp2p) that binds actin filament barbed ends. Neither acp1 nor acp2 is required for viability, but cells lacking either or both subunits have cytokinesis defects under stressful conditions, including elevated temperature, osmotic stress, or in combination with numerous mild mutations in genes important for cytokinesis. Defects arise as the contractile ring constricts and disassembles, resulting in delays in cell separation. Genetic and biochemical interactions show that the cytokinesis formin Cdc12p competes with capping protein for actin filament barbed ends in cells. Deletion of acp2 partly suppresses cytokinesis defects in temperature-sensitive cdc12-112 cells and mild overexpression of capping protein kills cdc12-112 cells. Biochemically, profilin has opposite effects on filaments capped with Cdc12p and capping protein. Profilin depolymerizes actin filaments capped by capping protein but allows filaments capped by Cdc12p to grow at their barbed ends. Once associated with a barbed end, either Cdc12p or capping protein prevents the other from influencing polymerization at that end. Given that capping protein arrives at the division site 20 min later than Cdc12p, capping protein may slowly replace Cdc12p on filament barbed ends in preparation for filament disassembly during ring constriction.
Figures




References
-
- Amatruda, J. F., Cannon, J. F., Tatchell, K., Hug, C., and Cooper, J. A. (1990). Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature 344, 352–354. - PubMed
-
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

