GIL16, a new gram-positive tectiviral phage related to the Bacillus thuringiensis GIL01 and the Bacillus cereus pBClin15 elements
- PMID: 15743944
- PMCID: PMC1064052
- DOI: 10.1128/JB.187.6.1966-1973.2005
GIL16, a new gram-positive tectiviral phage related to the Bacillus thuringiensis GIL01 and the Bacillus cereus pBClin15 elements
Abstract
One of the most notable characteristics of Tectiviridae resides in their double-layer coats: the double-stranded DNA is located within a flexible lipoprotein vesicle covered by a rigid protein capsid. Despite their apparent rarity, tectiviruses have an extremely wide distribution compared to other phage groups. Members of this family have been found to infect gram-negative (PRD1 and relatives) as well as gram-positive (Bam35, GIL01, AP50, and phiNS11) hosts. Several reports have shown that tectiviruses infecting gram-negative bacteria are closely related, whereas no information is currently available on the genetic relationship among those infecting gram-positive bacteria. The present study reports the sequence of GIL16, a new isolate originating from Bacillus thuringiensis, and a genetic comparison of this isolate with the tectiviral bacteriophages Bam35 and GIL01, which originated from B. thuringiensis serovars Alesti and Israelensis, respectively. In contrast to PRD1 and its relatives, these are temperate bacteriophages existing as autonomous linear prophages within the host cell. Mutations in a particular motif in both the GIL01 and GIL16 phages are also shown to correlate with a switch to the lytic cycle. Interestingly, both bacterial viruses displayed narrow, yet slightly different, host spectrums. We also explore the hypothesis that pBClin15, a linear plasmid hosted by the Bacillus cereus reference strain ATCC 14579, is also a prophage. Sequencing of its inverted repeats at both extremities and a comparison with GIL01 and GIL16 emphasize its relationship to the Tectiviridae.
Figures
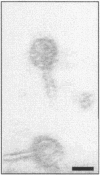



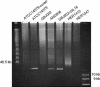
Similar articles
-
Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future.Viruses. 2014 Jul 9;6(7):2623-72. doi: 10.3390/v6072623. Viruses. 2014. PMID: 25010767 Free PMC article. Review.
-
pGIL01, a linear tectiviral plasmid prophage originating from Bacillus thuringiensis serovar israelensis.Microbiology (Reading). 2003 Aug;149(Pt 8):2083-2092. doi: 10.1099/mic.0.26307-0. Microbiology (Reading). 2003. PMID: 12904548
-
Prevalence, genetic diversity, and host range of tectiviruses among members of the Bacillus cereus group.Appl Environ Microbiol. 2014 Jul;80(14):4138-52. doi: 10.1128/AEM.00912-14. Epub 2014 May 2. Appl Environ Microbiol. 2014. PMID: 24795369 Free PMC article.
-
A snapshot of viral evolution from genome analysis of the tectiviridae family.J Mol Biol. 2005 Jul 15;350(3):427-40. doi: 10.1016/j.jmb.2005.04.059. J Mol Biol. 2005. PMID: 15946683
-
Characterization and comparative genomic analysis of bacteriophages infecting members of the Bacillus cereus group.Arch Virol. 2014 May;159(5):871-84. doi: 10.1007/s00705-013-1920-3. Epub 2013 Nov 22. Arch Virol. 2014. PMID: 24264384 Review.
Cited by
-
Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium.Genome Biol. 2012 Apr 26;13(4):R30. doi: 10.1186/gb-2012-13-4-r30. Genome Biol. 2012. PMID: 22537947 Free PMC article.
-
Comparative Genomics of Prophages Sato and Sole Expands the Genetic Diversity Found in the Genus Betatectivirus.Microorganisms. 2021 Jun 19;9(6):1335. doi: 10.3390/microorganisms9061335. Microorganisms. 2021. PMID: 34205474 Free PMC article.
-
Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future.Viruses. 2014 Jul 9;6(7):2623-72. doi: 10.3390/v6072623. Viruses. 2014. PMID: 25010767 Free PMC article. Review.
-
Disclosing early steps of protein-primed genome replication of the Gram-positive tectivirus Bam35.Nucleic Acids Res. 2016 Nov 16;44(20):9733-9744. doi: 10.1093/nar/gkw673. Epub 2016 Jul 27. Nucleic Acids Res. 2016. PMID: 27466389 Free PMC article.
-
Comparative transcriptomic and phenotypic analysis of the responses of Bacillus cereus to various disinfectant treatments.Appl Environ Microbiol. 2010 May;76(10):3352-60. doi: 10.1128/AEM.03003-09. Epub 2010 Mar 26. Appl Environ Microbiol. 2010. PMID: 20348290 Free PMC article.
References
-
- Ackermann, H. W., R. Roy, M. Martin, M. R. Murthy, and W. A. Smirnoff. 1978. Partial characterization of a cubic Bacillus phage. Can. J. Microbiol. 24:986-993. - PubMed
-
- Bamford, D. H., L. Rouhiainen, K. Takkinen, and H. Soderlund. 1981. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J. Gen. Virol. 57:365-373. - PubMed
-
- Bradley, D. E., and E. L. Rutherford. 1975. Basic characterization of a lipid-containing bacteriophage specific for plasmids of the P, N, and W compatibility groups. Can. J. Microbiol. 21:152-163. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

