A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6
- PMID: 15743951
- PMCID: PMC1064056
- DOI: 10.1128/JB.187.6.2030-2037.2005
A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6
Abstract
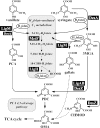
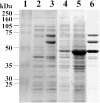
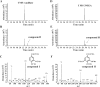
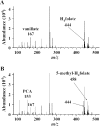
Vanillate and syringate are converted into protocatechuate (PCA) and 3-O-methylgallate (3MGA), respectively, by O-demethylases in Sphingomonas paucimobilis SYK-6. PCA is further degraded via the PCA 4,5-cleavage pathway, while 3MGA is degraded through multiple pathways in which PCA 4,5-dioxygenase (LigAB), 3MGA 3,4-dioxygenase (DesZ), and an unidentified 3MGA O-demethylase and gallate dioxygenase are participants. For this study, we isolated a 4.7-kb SmaI fragment that conferred on Escherichia coli the activity required for the conversion of vanillate to PCA. The nucleotide sequence of this fragment revealed an open reading frame of 1,413 bp (ligM), the deduced amino acid sequence of which showed 49% identity with that of the tetrahydrofolate (H4folate)-dependent syringate O-demethylase gene (desA). The metF and ligH genes, which are thought to be involved in H4folate-mediated C1 metabolism, were located just downstream of ligM. The crude LigM enzyme expressed in E. coli converted vanillate and 3MGA to PCA and gallate, respectively, with similar specific activities, and only in the presence of H4folate; however, syringate was not a substrate for LigM. The disruption of ligM led to significant growth retardation on both vanillate and syringate, indicating that ligM is involved in the catabolism of these substrates. The ability of the ligM mutant to transform vanillate was markedly decreased, and this mutant completely lost the 3MGA O-demethylase activity. A ligM desA double mutant completely lost the ability to transform vanillate, thus indicating that desA also contributes to vanillate degradation. All of these results indicate that ligM encodes vanillate/3MGA O-demethylase and plays an important role in the O demethylation of vanillate and 3MGA, respectively.
Figures

Similar articles
-
Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6.J Bacteriol. 2005 Aug;187(15):5067-74. doi: 10.1128/JB.187.15.5067-5074.2005. J Bacteriol. 2005. PMID: 16030198 Free PMC article.
-
Characterization of the 3-O-methylgallate dioxygenase gene and evidence of multiple 3-O-methylgallate catabolic pathways in Sphingomonas paucimobilis SYK-6.J Bacteriol. 2004 Aug;186(15):4951-9. doi: 10.1128/JB.186.15.4951-4959.2004. J Bacteriol. 2004. PMID: 15262932 Free PMC article.
-
A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate.J Bacteriol. 2004 May;186(9):2757-65. doi: 10.1128/JB.186.9.2757-2765.2004. J Bacteriol. 2004. PMID: 15090517 Free PMC article.
-
Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds.Biosci Biotechnol Biochem. 2007 Jan;71(1):1-15. doi: 10.1271/bbb.60437. Epub 2007 Jan 7. Biosci Biotechnol Biochem. 2007. PMID: 17213657 Review.
-
Microbial demethylation of lignin: Evidence of enzymes participating in the removal of methyl/methoxyl groups.Enzyme Microb Technol. 2021 Jun;147:109780. doi: 10.1016/j.enzmictec.2021.109780. Epub 2021 Mar 18. Enzyme Microb Technol. 2021. PMID: 33992403 Review.
Cited by
-
Transcriptional regulation of the terephthalate catabolism operon in Comamonas sp. strain E6.Appl Environ Microbiol. 2010 Sep;76(18):6047-55. doi: 10.1128/AEM.00742-10. Epub 2010 Jul 23. Appl Environ Microbiol. 2010. PMID: 20656871 Free PMC article.
-
Genomics and biochemistry investigation on the metabolic pathway of milled wood and alkali lignin-derived aromatic metabolites of Comamonas serinivorans SP-35.Biotechnol Biofuels. 2018 Dec 27;11:338. doi: 10.1186/s13068-018-1341-3. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30603046 Free PMC article.
-
Catabolic System of Syringic Acid, a Key Intermediate of Lignin-Derived Aromatic Compounds, via a Novel Linear Pathway in Pseudomonas sp. NGC7.J Agric Food Chem. 2025 Jul 30;73(30):18899-18913. doi: 10.1021/acs.jafc.5c04544. Epub 2025 Jul 16. J Agric Food Chem. 2025. PMID: 40668107 Free PMC article.
-
Kinetic modeling of anaerobic degradation of plant-derived aromatic mixtures by Rhodopseudomonas palustris.Biodegradation. 2021 Apr;32(2):179-192. doi: 10.1007/s10532-021-09932-3. Epub 2021 Mar 6. Biodegradation. 2021. PMID: 33675449 Free PMC article.
-
In Situ Wood Fiber Dyeing Through Laccase Catalysis for Fiberboard Production.Front Bioeng Biotechnol. 2021 Dec 3;9:778971. doi: 10.3389/fbioe.2021.778971. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34926424 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

