Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus
- PMID: 15743959
- PMCID: PMC1064035
- DOI: 10.1128/JB.187.6.2105-2112.2005
Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus
Abstract
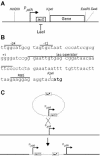
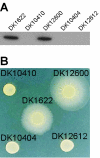
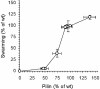
An isopropyl-beta-D-thiogalactopyranoside (IPTG)-inducible promoter was constructed in Myxococcus xanthus. The single-copy pilA gene encodes pilin, the monomer unit of M. xanthus type IV pili. To vary the level of pilA expression, we cloned its promoter in front of the lac operator, and a plasmid containing the construct was inserted into the chromosome of a DeltapilA strain. Induction of pilin expression increased smoothly as the dose of IPTG added to the culture was increased. IPTG-induced pilin rescued S motility of the DeltapilA strain to wild-type levels. The rate of S-motile swarming was found to be proportional to the number of pili (shear-sensitive pilin) produced rather than to the level of total pilin. In fact, S motility was not rescued until the total level of pilin was more than 50% of the wild-type level. This observation implies that a threshold concentration of pilin must be exceeded before the shear-sensitive material (pili) is polymerized in M. xanthus.
Figures





Similar articles
-
Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili.Microbiology (Reading). 2005 Feb;151(Pt 2):353-360. doi: 10.1099/mic.0.27614-0. Microbiology (Reading). 2005. PMID: 15699186
-
Characterization of four type IV pilin homologues in Stigmatella aurantiaca DSM17044 by heterologous expression in Myxococcus xanthus.PLoS One. 2013 Sep 18;8(9):e75105. doi: 10.1371/journal.pone.0075105. eCollection 2013. PLoS One. 2013. PMID: 24058653 Free PMC article.
-
Alanine 32 in PilA is important for PilA stability and type IV pili function in Myxococcus xanthus.Microbiology (Reading). 2011 Jul;157(Pt 7):1920-1928. doi: 10.1099/mic.0.049684-0. Epub 2011 Apr 14. Microbiology (Reading). 2011. PMID: 21493683 Free PMC article.
-
Gliding motility in bacteria: insights from studies of Myxococcus xanthus.Microbiol Mol Biol Rev. 1999 Sep;63(3):621-41. doi: 10.1128/MMBR.63.3.621-641.1999. Microbiol Mol Biol Rev. 1999. PMID: 10477310 Free PMC article. Review.
-
Regulation of Cell Polarity in Motility and Cell Division in Myxococcus xanthus.Annu Rev Microbiol. 2017 Sep 8;71:61-78. doi: 10.1146/annurev-micro-102215-095415. Epub 2017 May 19. Annu Rev Microbiol. 2017. PMID: 28525300 Review.
Cited by
-
Two systems for conditional gene expression in Myxococcus xanthus inducible by isopropyl-β-D-thiogalactopyranoside or vanillate.J Bacteriol. 2012 Nov;194(21):5875-85. doi: 10.1128/JB.01110-12. Epub 2012 Aug 24. J Bacteriol. 2012. PMID: 22923595 Free PMC article.
-
Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa.J Bacteriol. 2010 Feb;192(4):994-1010. doi: 10.1128/JB.01390-09. Epub 2009 Dec 11. J Bacteriol. 2010. PMID: 20008072 Free PMC article.
-
PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus.J Bacteriol. 2008 Apr;190(7):2411-21. doi: 10.1128/JB.01793-07. Epub 2008 Jan 25. J Bacteriol. 2008. PMID: 18223089 Free PMC article.
-
Myxococcus xanthus PilB interacts with c-di-GMP and modulates motility and biofilm formation.J Bacteriol. 2023 Sep 26;205(9):e0022123. doi: 10.1128/jb.00221-23. Epub 2023 Sep 11. J Bacteriol. 2023. PMID: 37695853 Free PMC article.
-
The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria.PLoS One. 2008 May 7;3(5):e2103. doi: 10.1371/journal.pone.0002103. PLoS One. 2008. PMID: 18461135 Free PMC article.
References
-
- Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in M. xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176.
-
- Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in M. xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

