Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis
- PMID: 15743967
- PMCID: PMC1064044
- DOI: 10.1128/JB.187.6.2182-2189.2005
Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis
Abstract
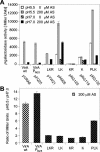
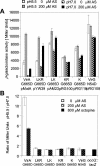
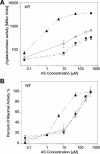
Agrobacterium tumefaciens stands as one of biotechnology's greatest successes, with all plant genetic engineering building on the strategies of this pathogen. By integrating responses to external pHs, phenols, and monosaccharides, this organism mobilizes oncogenic elements to efficiently transform most dicotyledonous plants. We now show that the complex signaling network used to regulate lateral gene transfer can be resolved as individual signaling modules. While pH and sugar perception are coupled through a common pathway, requiring both low pH and sugar for maximal virulence gene expression, various VirA and ChvE alleles can decouple pH and monosaccharide perception. This VirA and ChvE system may represent a common mechanism that underpins external pH perception in prokaryotes, and the use of these simple genetic elements may now be extended to research on specific responses to changes in environmental pH.
Figures


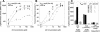

Similar articles
-
Reconstitution of acetosyringone-mediated Agrobacterium tumefaciens virulence gene expression in the heterologous host Escherichia coli.J Bacteriol. 2001 Jun;183(12):3704-11. doi: 10.1128/JB.183.12.3704-3711.2001. J Bacteriol. 2001. PMID: 11371534 Free PMC article.
-
Glu-255 outside the predicted ChvE binding site in VirA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens.J Bacteriol. 1994 Jun;176(11):3242-9. doi: 10.1128/jb.176.11.3242-3249.1994. J Bacteriol. 1994. PMID: 8195079 Free PMC article.
-
Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein.J Bacteriol. 1992 Nov;174(21):7033-9. doi: 10.1128/jb.174.21.7033-7039.1992. J Bacteriol. 1992. PMID: 1400253 Free PMC article.
-
Capturing the VirA/VirG TCS of Agrobacterium tumefaciens.Adv Exp Med Biol. 2008;631:161-77. doi: 10.1007/978-0-387-78885-2_11. Adv Exp Med Biol. 2008. PMID: 18792688 Review.
-
Host recognition by the VirA, VirG two-component regulatory proteins of agrobacterium tumefaciens.Res Microbiol. 1994 Jun-Aug;145(5-6):461-73. doi: 10.1016/0923-2508(94)90095-7. Res Microbiol. 1994. PMID: 7855433 Review.
Cited by
-
Evolutionary tuning of protein expression levels of a positively autoregulated two-component system.PLoS Genet. 2013 Oct;9(10):e1003927. doi: 10.1371/journal.pgen.1003927. Epub 2013 Oct 24. PLoS Genet. 2013. PMID: 24204322 Free PMC article.
-
Agrobacterium tumefaciens responses to plant-derived signaling molecules.Front Plant Sci. 2014 Jul 8;5:322. doi: 10.3389/fpls.2014.00322. eCollection 2014. Front Plant Sci. 2014. PMID: 25071805 Free PMC article. Review.
-
The Agrobacterium Ti Plasmids.Microbiol Spectr. 2014 Dec;2(6):10.1128/microbiolspec.PLAS-0010-2013. doi: 10.1128/microbiolspec.PLAS-0010-2013. Microbiol Spectr. 2014. PMID: 25593788 Free PMC article. Review.
-
Synergistic Action of D-Glucose and Acetosyringone on Agrobacterium Strains for Efficient Dunaliella Transformation.PLoS One. 2016 Jun 28;11(6):e0158322. doi: 10.1371/journal.pone.0158322. eCollection 2016. PLoS One. 2016. PMID: 27351975 Free PMC article.
-
Constitutive activation of two-component response regulators: characterization of VirG activation in Agrobacterium tumefaciens.J Bacteriol. 2006 Jul;188(14):5204-11. doi: 10.1128/JB.00387-06. J Bacteriol. 2006. PMID: 16816192 Free PMC article.
References
-
- Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

