Maternal transmission ratio distortion at the mouse Om locus results from meiotic drive at the second meiotic division
- PMID: 15744049
- PMCID: PMC1449735
- DOI: 10.1534/genetics.104.039479
Maternal transmission ratio distortion at the mouse Om locus results from meiotic drive at the second meiotic division
Abstract
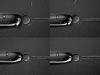
We have observed maternal transmission ratio distortion (TRD) in favor of DDK alleles at the Ovum mutant (Om) locus on mouse chromosome 11 among the offspring of (C57BL/6 x DDK) F(1) females and C57BL/6 males. Although significant lethality occurs in this backcross ( approximately 50%), differences in the level of TRD found in recombinant vs. nonrecombinant chromosomes among offspring argue that TRD is due to nonrandom segregation of chromatids at the second meiotic division, i.e., true meiotic drive. We tested this hypothesis directly, by determining the centromere and Om genotypes of individual chromatids in zygote stage embryos. We found similar levels of TRD in favor of DDK alleles at Om in the female pronucleus and TRD in favor of C57BL/6 alleles at Om in the second polar body. In those embryos for which complete dyads have been reconstructed, TRD was present only in those inheriting heteromorphic dyads. These results demonstrate that meiotic drive occurs at MII and that preferential death of one genotypic class of embryo does not play a large role in the TRD.
Figures

Similar articles
-
A genetic test to determine the origin of maternal transmission ratio distortion. Meiotic drive at the mouse Om locus.Genetics. 2000 Jan;154(1):333-42. doi: 10.1093/genetics/154.1.333. Genetics. 2000. PMID: 10628992 Free PMC article.
-
Heritability of the maternal meiotic drive system linked to Om and high-resolution mapping of the Responder locus in mouse.Genetics. 2000 May;155(1):283-9. doi: 10.1093/genetics/155.1.283. Genetics. 2000. PMID: 10790402 Free PMC article.
-
Male-offspring-specific, haplotype-dependent, nonrandom cosegregation of alleles at loci on two mouse chromosomes.Genetics. 2000 Jan;154(1):351-6. doi: 10.1093/genetics/154.1.351. Genetics. 2000. PMID: 10628994 Free PMC article.
-
Non-Mendelian segregation and transmission drive of B chromosomes.Chromosome Res. 2022 Sep;30(2-3):217-228. doi: 10.1007/s10577-022-09692-7. Epub 2022 Jun 3. Chromosome Res. 2022. PMID: 35657532 Review.
-
Src protein kinases in mouse and rat oocytes and embryos.Results Probl Cell Differ. 2012;55:93-106. doi: 10.1007/978-3-642-30406-4_5. Results Probl Cell Differ. 2012. PMID: 22918802 Review.
Cited by
-
Germ cell nuclear factor regulates gametogenesis in developing gonads.PLoS One. 2014 Aug 20;9(8):e103985. doi: 10.1371/journal.pone.0103985. eCollection 2014. PLoS One. 2014. PMID: 25140725 Free PMC article.
-
Scrambling eggs: meiotic drive and the evolution of female recombination rates.Genetics. 2012 Feb;190(2):709-23. doi: 10.1534/genetics.111.136721. Epub 2011 Dec 5. Genetics. 2012. PMID: 22143919 Free PMC article.
-
A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver's antidote.PLoS Genet. 2018 Nov 26;14(11):e1007836. doi: 10.1371/journal.pgen.1007836. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30475921 Free PMC article.
-
Bypassing Mendel's First Law: Transmission Ratio Distortion in Mammals.Int J Mol Sci. 2023 Jan 13;24(2):1600. doi: 10.3390/ijms24021600. Int J Mol Sci. 2023. PMID: 36675116 Free PMC article. Review.
-
Role of glucose in cloned mouse embryo development.Am J Physiol Endocrinol Metab. 2008 Oct;295(4):E798-809. doi: 10.1152/ajpendo.00683.2007. Epub 2008 Jun 24. Am J Physiol Endocrinol Metab. 2008. PMID: 18577693 Free PMC article.
References
-
- Agulnik, S. I., A. I. Agulnik and A. O. Ruvinsky, 1990. Meiotic drive in female mice heterozygous for the HSR inserts on chromosome 1. Genet. Res. 55: 97–100. - PubMed
-
- Babinet, C., V. Richoux, J. L. Guenet and J. P. Renard, 1990 The DDK inbred strain as a model for the study of interactions between parental genomes and egg cytoplasm in mouse preimplantation development. Development S: 81–87. - PubMed
-
- Baldacci, P. A., M. Cohen-Tannoudji, C. Kress, S. Pournin and C. Babinet, 1996. A high-resolution map around the locus Om on mouse chromosome 11. Mamm. Genome 7: 114–116. - PubMed
-
- Chatot, C. L., C. A. Ziomek, B. D. Bavister, J. L. Lewis and I. Torres, 1989. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 86: 679–688. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

