Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule
- PMID: 15744050
- PMCID: PMC1449729
- DOI: 10.1534/genetics.104.039396
Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule
Abstract
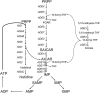
Because some metabolic intermediates are involved in more than one pathway, crosstalk between pathways is crucial to maintaining homeostasis. AMP and histidine biosynthesis pathways are coregulated at the transcriptional level in response to adenine availability. 5'-Phosphoribosyl-4-carboxamide-5-aminoimidazole (AICAR), a metabolic intermediate at the crossroads between these two pathways, is shown here to be critical for activation of the transcriptional response in the absence of adenine. In this study, we show that both AMP and histidine pathways significantly contribute to AICAR synthesis. Furthermore, we show that upregulation of the histidine pathway clearly interferes with regulation of the AMP pathway, thus providing an explanation for the regulatory crosstalk between these pathways. Finally, we revisit the histidine auxotrophy of ade3 or ade16 ade17 mutants. Interestingly, overexpression of PMU1, encoding a potential phosphomutase, partially suppresses the histidine requirement of an ade3 ade16 ade17 triple mutant, most probably by reducing the level of AICAR in this mutant. Together our data clearly establish that AICAR is not just a metabolic intermediate but also acts as a true regulatory molecule.
Figures
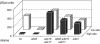





Similar articles
-
Serine hydroxymethyltransferase: a key player connecting purine, folate and methionine metabolism in Saccharomyces cerevisiae.Curr Genet. 2015 Nov;61(4):633-40. doi: 10.1007/s00294-015-0489-7. Epub 2015 Apr 17. Curr Genet. 2015. PMID: 25893566
-
Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate.Mol Cell Biol. 2001 Dec;21(23):7901-12. doi: 10.1128/MCB.21.23.7901-7912.2001. Mol Cell Biol. 2001. PMID: 11689683 Free PMC article.
-
DNA-bound Bas1 recruits Pho2 to activate ADE genes in Saccharomyces cerevisiae.Eukaryot Cell. 2005 Oct;4(10):1725-35. doi: 10.1128/EC.4.10.1725-1735.2005. Eukaryot Cell. 2005. PMID: 16215179 Free PMC article.
-
Purine Chemistry in the Early RNA World at the Origins of Life: From RNA and Nucleobases Lesions to Current Key Metabolic Routes.Chembiochem. 2025 Jun 3;26(11):e202500035. doi: 10.1002/cbic.202500035. Epub 2025 Apr 16. Chembiochem. 2025. PMID: 40237374 Free PMC article. Review.
-
Biochemical strategy of cancer cells and the design of chemotherapy: G. H. A. Clowes Memorial Lecture.Cancer Res. 1983 Aug;43(8):3466-92. Cancer Res. 1983. PMID: 6305486 Review. No abstract available.
Cited by
-
Purine metabolism in plant pathogenic fungi.Front Microbiol. 2024 Feb 7;15:1352354. doi: 10.3389/fmicb.2024.1352354. eCollection 2024. Front Microbiol. 2024. PMID: 38384269 Free PMC article. Review.
-
Metabolic and physiological responses to progressive drought stress in bread wheat.Sci Rep. 2020 Oct 14;10(1):17189. doi: 10.1038/s41598-020-74303-6. Sci Rep. 2020. PMID: 33057205 Free PMC article.
-
Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae.Genetics. 2012 Mar;190(3):885-929. doi: 10.1534/genetics.111.133306. Genetics. 2012. PMID: 22419079 Free PMC article.
-
Quantitative analysis of Mycobacterium avium subsp. hominissuis proteome in response to antibiotics and during exposure to different environmental conditions.Clin Proteomics. 2019 Nov 13;16:39. doi: 10.1186/s12014-019-9260-2. eCollection 2019. Clin Proteomics. 2019. PMID: 31749666 Free PMC article.
-
Dissection of the PHO pathway in Schizosaccharomyces pombe using epistasis and the alternate repressor adenine.Curr Genet. 2015 May;61(2):175-83. doi: 10.1007/s00294-014-0466-6. Epub 2014 Dec 30. Curr Genet. 2015. PMID: 25547512
References
-
- Arndt, K. T., C. Styles and G. R. Fink, 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237: 874–880. - PubMed
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. - PubMed
-
- Corton, J. M., J. G. Gillespie, S. A. Hawley and D. G. Hardie, 1995. 5-Aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 229: 558–565. - PubMed
-
- Cross, F. R., 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13: 647–653. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

