Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist
- PMID: 15745995
- PMCID: PMC552970
- DOI: 10.1093/nar/gni044
Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist
Abstract
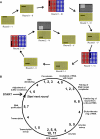
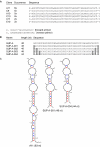
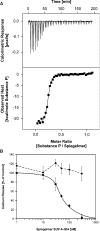
We have developed an automated SELEX (Systematic Evolution of Ligands by EXponential Enrichment) process that allows the execution of in vitro selection cycles without any direct manual intervention steps. The automated selection protocol is designed to provide for high flexibility and versatility in terms of choice of buffers and reagents as well as stringency of selection conditions. Employing the automated SELEX process, we have identified RNA aptamers to the mirror-image configuration (d-peptide) of substance P. The peptide substance P belongs to the tachykinin family and exerts various biologically important functions, such as peripheral vasodilation, smooth muscle contraction and pain transmission. The aptamer that was identified most frequently was truncated to the 44mer SUP-A-004. The mirror-image configuration of SUP-A-004, the so-called Spiegelmer, has been shown to bind to naturally occurring l-substance P displaying a K(d) of 40 nM and to inhibit (IC50 of 45 nM) l-substance P-mediated Ca2+ release in a cell culture assay.
Figures


Similar articles
-
In vitro selection using a dual RNA library that allows primerless selection.Nucleic Acids Res. 2006 Jul 19;34(12):e86. doi: 10.1093/nar/gkl463. Nucleic Acids Res. 2006. PMID: 16855281 Free PMC article.
-
Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface.Biochimie. 2006 Jul;88(7):897-904. doi: 10.1016/j.biochi.2006.02.004. Epub 2006 Mar 6. Biochimie. 2006. PMID: 16540230
-
Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers.RNA. 2007 Apr;13(4):614-22. doi: 10.1261/rna.334307. Epub 2007 Feb 5. RNA. 2007. PMID: 17283213 Free PMC article.
-
SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands.Biomol Eng. 2007 Oct;24(4):381-403. doi: 10.1016/j.bioeng.2007.06.001. Epub 2007 Jun 16. Biomol Eng. 2007. PMID: 17627883 Review.
-
Disease-specific biomarker discovery by aptamers.Cytometry A. 2009 Sep;75(9):727-33. doi: 10.1002/cyto.a.20766. Cytometry A. 2009. PMID: 19565638 Review.
Cited by
-
Polyetheylenimine-polyplexes of Spiegelmer NOX-A50 directed against intracellular high mobility group protein A1 (HMGA1) reduce tumor growth in vivo.J Biol Chem. 2010 Dec 17;285(51):40012-8. doi: 10.1074/jbc.M110.178533. Epub 2010 Oct 20. J Biol Chem. 2010. PMID: 20961861 Free PMC article.
-
Applications in Which Aptamers Are Needed or Wanted in Diagnostics and Therapeutics.Pharmaceuticals (Basel). 2022 Jun 1;15(6):693. doi: 10.3390/ph15060693. Pharmaceuticals (Basel). 2022. PMID: 35745612 Free PMC article. Review.
-
Aptamers in Virology-A Consolidated Review of the Most Recent Advancements in Diagnosis and Therapy.Pharmaceutics. 2021 Oct 9;13(10):1646. doi: 10.3390/pharmaceutics13101646. Pharmaceutics. 2021. PMID: 34683938 Free PMC article. Review.
-
Aptamer and its applications in neurodegenerative diseases.Cell Mol Life Sci. 2017 Feb;74(4):683-695. doi: 10.1007/s00018-016-2345-4. Epub 2016 Aug 25. Cell Mol Life Sci. 2017. PMID: 27563707 Free PMC article. Review.
-
Progress and Challenges in Developing Aptamer-Functionalized Targeted Drug Delivery Systems.Int J Mol Sci. 2015 Oct 9;16(10):23784-822. doi: 10.3390/ijms161023784. Int J Mol Sci. 2015. PMID: 26473828 Free PMC article. Review.
References
-
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. - PubMed
-
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
-
- Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. - PubMed
-
- Brody E.N., Gold L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000;74:5–13. - PubMed
-
- Cox J.C., Rudolph P., Ellington A.D. Automated RNA selection. Biotechnol. Prog. 1998;14:845–850. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

