Cadmium inhibits mismatch repair by blocking the ATPase activity of the MSH2-MSH6 complex
- PMID: 15746000
- PMCID: PMC552968
- DOI: 10.1093/nar/gki291
Cadmium inhibits mismatch repair by blocking the ATPase activity of the MSH2-MSH6 complex
Erratum in
- Nucleic Acids Res. 2005;33(5):1738
Abstract
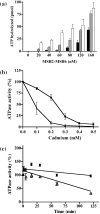
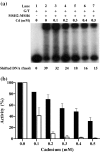
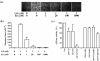
Cadmium (Cd2+) is a known carcinogen that inactivates the DNA mismatch repair (MMR) pathway. In this study, we have tested the effect of Cd2+ exposure on the enzymatic activity of the mismatch binding complex MSH2-MSH6. Our results indicate that Cd2+ is highly inhibitory to the ATP binding and hydrolysis activities of MSH2-MSH6, and less inhibitory to its DNA mismatch binding activity. The inhibition of the ATPase activity appears to be dose and exposure time dependent. However, the inhibition of the ATPase activity by Cd2+ is prevented by cysteine and histidine, suggesting that these residues are essential for the ATPase activity and are targeted by Cd2+. A comparison of the mechanism of inhibition with N-ethyl maleimide, a sulfhydryl group inhibitor, indicates that this inhibition does not occur through direct inactivation of sulfhydryl groups. Zinc (Zn2+) does not overcome the direct inhibitory effect of Cd2+ on the MSH2-MSH6 ATPase activity in vitro. However, the increase in the mutator phenotype of yeast cells exposed to Cd2+ was prevented by excess Zn2+, probably by blocking the entry of Cd2+ into the cell. We conclude that the inhibition of MMR by Cd2+ is through the inactivation of the ATPase activity of the MSH2-MSH6 heterodimer, resulting in a dominant negative effect and causing a mutator phenotype.
Figures


References
-
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. - PubMed
-
- Kolodner R.D., Marsischky G.T. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 1999;9:89–96. - PubMed
-
- Modrich P., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. - PubMed
-
- Buermeyer A.B., Deschenes S.M., Baker S.M., Liskay R.M. Mammalian DNA mismatch repair. Annu. Rev. Genet. 1999;33:533–564. - PubMed
-
- Drotschmann K., Yang W., Kunkel T.A. Evidence for sequential action of two ATPase active sites in yeast Msh2–Msh6. DNA Repair (Amst.) 2002;1:743–753. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

