Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5
- PMID: 15746341
- PMCID: PMC1065149
- DOI: 10.1128/AEM.71.3.1387-1393.2005
Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5
Abstract
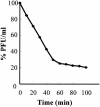
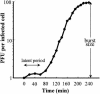
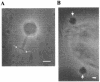
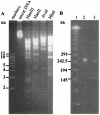
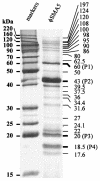
Stenotrophomonas maltophilia is one of the most prevalent opportunistic bacteria causing nosocomial infections. It has become problematic because most of the isolates are resistant to multiple antibiotics, and therefore, development of phage therapy has attracted strong attention. In this study, eight S. maltophilia phages were isolated from clinical samples including patient specimens, catheter-related devices, and wastewater. These phages can be divided into four distinct groups based on host range and digestibility of the phage DNAs with different restriction endonucleases. One of them, designated phiSMA5, was further characterized. Electron microscopy showed it resembled Myoviridae, with an isometric head (90 nm in diameter), a tail (90 nm long), a baseplate (25 nm wide), and short tail fibers. The phiSMA5 double-stranded DNA, refractory to digestion by most restriction enzymes, was tested and estimated to be 250 kb by pulsed-field gel electrophoresis. This genome size is second to that of the largest phage, phiKZ of Pseudomonas aeruginosa. In sodium dodecyl sulfate-polyacrylamide gel electrophoresis, 25 virion proteins were visualized. N-terminal sequencing of four of them suggested that each of them might have had its N terminus cleaved off. Among the 87 S. maltophilia strains collected in this study, only 61 were susceptible to phiSMA5, indicating that more phages are needed toward a phage therapy strategy. Since literature search yielded no information about S. maltophilia phages, phiSMA5 appears to be the first reported.
Figures
Similar articles
-
Genomic sequence of temperate phage Smp131 of Stenotrophomonas maltophilia that has similar prophages in xanthomonads.BMC Microbiol. 2014 Jan 28;14:17. doi: 10.1186/1471-2180-14-17. BMC Microbiol. 2014. PMID: 24472137 Free PMC article.
-
Characterization of a novel T4-type Stenotrophomonas maltophilia virulent phage Smp14.Arch Microbiol. 2007 Aug;188(2):191-7. doi: 10.1007/s00203-007-0238-5. Epub 2007 Apr 18. Arch Microbiol. 2007. PMID: 17440710
-
Isolation and characterization of a novel filamentous phage from Stenotrophomonas maltophilia.Arch Virol. 2012 Sep;157(9):1643-50. doi: 10.1007/s00705-012-1305-z. Epub 2012 May 22. Arch Virol. 2012. PMID: 22614810
-
The Potential of Phage Therapy against the Emerging Opportunistic Pathogen Stenotrophomonas maltophilia.Viruses. 2021 Jun 3;13(6):1057. doi: 10.3390/v13061057. Viruses. 2021. PMID: 34204897 Free PMC article. Review.
-
The SPO1-related bacteriophages.Arch Virol. 2010 Oct;155(10):1547-61. doi: 10.1007/s00705-010-0783-0. Epub 2010 Aug 17. Arch Virol. 2010. PMID: 20714761 Review.
Cited by
-
Enhancement of bactericidal effects of bacteriophage and gentamicin combination regimen against Staphylococcus aureus and Pseudomonas aeruginosa strains in a mice diabetic wound model.Virus Genes. 2024 Feb;60(1):80-96. doi: 10.1007/s11262-023-02037-4. Epub 2023 Dec 11. Virus Genes. 2024. PMID: 38079060
-
Characterization of Novel Broad-Host-Range Bacteriophage DLP3 Specific to Stenotrophomonas maltophilia as a Potential Therapeutic Agent.Front Microbiol. 2020 Jun 24;11:1358. doi: 10.3389/fmicb.2020.01358. eCollection 2020. Front Microbiol. 2020. PMID: 32670234 Free PMC article.
-
Complete Genome Sequence of a Novel T7-Like Bacteriophage from a Pasteurella multocida Capsular Type A Isolate.Curr Microbiol. 2018 May;75(5):574-579. doi: 10.1007/s00284-017-1419-3. Epub 2018 Jan 6. Curr Microbiol. 2018. PMID: 29307051
-
Genomic sequence of temperate phage Smp131 of Stenotrophomonas maltophilia that has similar prophages in xanthomonads.BMC Microbiol. 2014 Jan 28;14:17. doi: 10.1186/1471-2180-14-17. BMC Microbiol. 2014. PMID: 24472137 Free PMC article.
-
Complete Genome Sequence of Temperate Stenotrophomonas maltophilia Bacteriophage DLP5.Genome Announc. 2018 Mar 1;6(9):e00073-18. doi: 10.1128/genomeA.00073-18. Genome Announc. 2018. PMID: 29496826 Free PMC article.
References
-
- Ackermann, H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843-857. - PubMed
-
- Ackermann, H. W. 2003. Bacteriophage observations and evolution. Res. Microbiol. 154:245-251. - PubMed
-
- Aznar, R., E. Alcaide, and E. Garay. 1992. Numerical taxonomy of pseudomonads isolated from water, sediment and eels. Syst. Appl. Microbiol. 14:235-246.
-
- Ballestero, S., I. Virseda, H. Escobar, L. Suarez, and F. Baquero. 1995. Stenotrophomonas maltophilia in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 14:728-729. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

