Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations
- PMID: 15746368
- PMCID: PMC1065155
- DOI: 10.1128/AEM.71.3.1626-1637.2005
Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations
Abstract
Microorganisms, such as bacteria, which might be present as contamination inside an industrial food or pharmaceutical clean room process need to be identified on short time scales in order to minimize possible health hazards as well as production downtimes causing financial deficits. Here we describe the first results of single-particle micro-Raman measurements in combination with a classification method, the so-called support vector machine technique, allowing for a fast, reliable, and nondestructive online identification method for single bacteria.
Figures


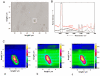

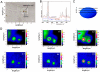


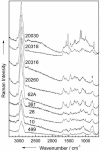
References
-
- Alexander, T. A., P. M. Pellegrino, and J. B. Gillespie. 2003. Near-infrared surface-enhanced-Raman-scattering (SERS) mediated identification of single optically trapped bacterial spores. Proc. SPIE 5085:91-100. - PubMed
-
- Al-Khaldi, S. F., and M. M. Mossoba. 2004. Gene and bacterial identification using high-throughput technologies: genomics, proteomics, and phenomics. Nutrition 20:32-38. - PubMed
-
- Berger, A. J., and Q. Zhu. 2003. Identification of oral bacteria by Raman microspectroscopy. J. Mod. Optics 50:2375-2380.
-
- Britton, K. A., R. A. Dalterio, W. H. Nelson, D. Britt, and J. F. Sperry. 1988. Ultraviolet resonance Raman spectra of Escherichia coli with 222.5-251.0 nm pulsed laser excitation. Appl. Spectrosc. 42:782-788.
-
- Burges, C. J. C. 1998. A tutorial on support vector machines for pattern recognition. Data Mining Knowledge Disc. 2:121-167.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

