A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms
- PMID: 15747258
- PMCID: PMC1199300
- DOI: 10.1086/429393
A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms
Abstract
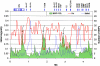
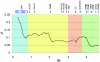
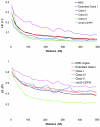
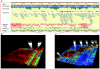
Autoimmune, inflammatory, and infectious diseases present a major burden to human health and are frequently associated with loci in the human major histocompatibility complex (MHC). Here, we report a high-resolution (1.9 kb) linkage-disequilibrium (LD) map of a 4.46-Mb fragment containing the MHC in U.S. pedigrees with northern and western European ancestry collected by the Centre d'Etude du Polymorphisme Humain (CEPH) and the first generation of haplotype tag single-nucleotide polymorphisms (tagSNPs) that provide up to a fivefold increase in genotyping efficiency for all future MHC-linked disease-association studies. The data confirm previously identified recombination hotspots in the class II region and allow the prediction of numerous novel hotspots in the class I and class III regions. The region of longest LD maps outside the classic MHC to the extended class I region spanning the MHC-linked olfactory-receptor gene cluster. The extended haplotype homozygosity analysis for recent positive selection shows that all 14 outlying haplotype variants map to a single extended haplotype, which most commonly bears HLA-DRB1*1501. The SNP data, haplotype blocks, and tagSNPs analysis reported here have been entered into a multidimensional Web-based database (GLOVAR), where they can be accessed and viewed in the context of relevant genome annotation. This LD map allowed us to give coordinates for the extremely variable LD structure underlying the MHC.
Figures


References
Electronic-Database Information
-
- Center for Statistical Genetics, http://www.sph.umich.edu/csg/abecasis/PedStats/ (for PEDSTATS)
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html
-
- GLOVAR Genome Browser, http://www.glovar.org/Homo_sapiens/
-
- Human Chromosome 6 Project Overview, http://www.sanger.ac.uk/HGP/Chr6/
References
-
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) MERLIN—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 - PubMed
-
- Abecasis GR, Cookson WOC (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 - PubMed
-
- Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling K-L, Barnardo M, Goldthorpe S, Walton R, Bunce M, Jewell DP, Welsh KI (2003) Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet 12:647–656 - PubMed
-
- Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, Swerdlin A, Perick-Vance MA, Haines JL, Hauser SL, for the Multiple Sclerosis Genetics Group (2003) HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet 72:710–716 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

