Dissecting the genetics of human high myopia: a molecular biologic approach
- PMID: 15747770
- PMCID: PMC1280112
Dissecting the genetics of human high myopia: a molecular biologic approach
Abstract
Purpose: Despite the plethora of experimental myopia animal studies that demonstrate biochemical factor changes in various eye tissues, and limited human studies utilizing pharmacologic agents to thwart axial elongation, we have little knowledge of the basic physiology that drives myopic development. Identifying the implicated genes for myopia susceptibility will provide a fundamental molecular understanding of how myopia occurs and may lead to directed physiologic (ie, pharmacologic, gene therapy) interventions. The purpose of this proposal is to describe the results of positional candidate gene screening of selected genes within the autosomal dominant high-grade myopia-2 locus (MYP2) on chromosome 18p11.31.
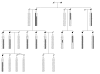
Methods: A physical map of a contracted MYP2 interval was compiled, and gene expression studies in ocular tissues using complementary DNA library screens, microarray matches, and reverse-transcription techniques aided in prioritizing gene selection for screening. The TGIF, EMLIN-2, MLCB, and CLUL1 genes were screened in DNA samples from unrelated controls and in high-myopia affected and unaffected family members from the original seven MYP2 pedigrees. All candidate genes were screened by direct base pair sequence analysis.
Results: Consistent segregation of a gene sequence alteration (polymorphism) with myopia was not demonstrated in any of the seven families. Novel single nucleotide polymorphisms were found.
Conclusion: The positional candidate genes TGIF, EMLIN-2, MLCB, and CLUL1 are not associated with MYP2-linked high-grade myopia. Base change polymorphisms discovered with base sequence screening of these genes were submitted to an Internet database. Other genes that also map within the interval are currently undergoing mutation screening.
Figures




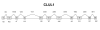
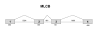

Similar articles
-
Insight into the molecular genetics of myopia.Mol Vis. 2017 Dec 31;23:1048-1080. eCollection 2017. Mol Vis. 2017. PMID: 29386878 Free PMC article. Review.
-
Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes.Mol Vis. 2005 Feb 2;11:97-110. Mol Vis. 2005. PMID: 15723005
-
Sequence variants in the transforming growth beta-induced factor (TGIF) gene are not associated with high myopia.Invest Ophthalmol Vis Sci. 2004 Jul;45(7):2091-7. doi: 10.1167/iovs.03-0933. Invest Ophthalmol Vis Sci. 2004. PMID: 15223781
-
Evaluation of Lipin 2 as a candidate gene for autosomal dominant 1 high-grade myopia.Gene. 2005 Jun 6;352:10-9. doi: 10.1016/j.gene.2005.02.019. Gene. 2005. PMID: 15862761
-
Further refinement of the MYP2 locus for autosomal dominant high myopia by linkage disequilibrium analysis.Ophthalmic Genet. 2001 Jun;22(2):69-75. doi: 10.1076/opge.22.2.69.2233. Ophthalmic Genet. 2001. PMID: 11449316
Cited by
-
Severe anisometropic myopia in identical twins.Middle East Afr J Ophthalmol. 2014 Jul-Sep;21(3):275-8. doi: 10.4103/0974-9233.134695. Middle East Afr J Ophthalmol. 2014. PMID: 25100917 Free PMC article.
-
Anti-VEGF treatment for myopic choroid neovascularization: from molecular characterization to update on clinical application.Drug Des Devel Ther. 2015 Jul 2;9:3413-21. doi: 10.2147/DDDT.S87920. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26170626 Free PMC article. Review.
-
Update on Myopia Risk Factors and Microenvironmental Changes.J Ophthalmol. 2019 Oct 31;2019:4960852. doi: 10.1155/2019/4960852. eCollection 2019. J Ophthalmol. 2019. PMID: 31781378 Free PMC article. Review.
-
Insight into the molecular genetics of myopia.Mol Vis. 2017 Dec 31;23:1048-1080. eCollection 2017. Mol Vis. 2017. PMID: 29386878 Free PMC article. Review.
-
Hepatocyte growth factor and myopia: genetic association analyses in a Caucasian population.Mol Vis. 2009 May 20;15:1028-35. Mol Vis. 2009. PMID: 19471602 Free PMC article.
References
-
- Curtin BJ. The Myopias: Basic Science and Clinical Management. New York: Harper & Rowe; 1985:237–245.
-
- Sperduto RD, Siegel D, Roberts J, et al. Prevalence of myopia in the United States. Arch Ophthalmol. 1983;101:405–407. - PubMed
-
- Wang Q, Klein, BEK, Klein R, et al. Refractive status in the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 1994;35:4344–4347. - PubMed
-
- Sperduto RD, Siegel D, Roberts J, et al. Prevalence of myopia in the United States. Arch Ophthalmol. 1983;101:405–407. - PubMed
-
- Angle J, Wissmann DA. The epidemiology of myopia. Am J Epidemiol. 1980;111:220–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
