Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV)
- PMID: 15749129
- PMCID: PMC7111783
- DOI: 10.1016/j.virol.2005.01.035
Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV)
Erratum in
- Virology. 2005 Jun 20;337(1):204. Mboudoudjeck, Innocent [corrected to Mboudjeka, Innocent]
Abstract
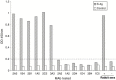
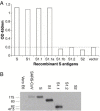
Inactivated severe acute respiratory syndrome-associated coronavirus (SARS-CoV) has been tested as a candidate vaccine against the re-emergence of SARS. In order to understand the efficacy and safety of this approach, it is important to know the antibody specificities generated with inactivated SARS-CoV. In the current study, a panel of twelve monoclonal antibodies (mAbs) was established by immunizing Balb/c mice with the inactivated BJ01 strain of SARS-CoV isolated from the lung tissue of a SARS-infected Chinese patient. These mAbs could recognize SARS-CoV-infected cells by immunofluorescence analysis (IFA). Seven of them were mapped to the specific segments of recombinant spike (S) protein: six on S1 subunit (aa 12-798) and one on S2 subunit (aa 797-1192). High neutralizing titers against SARS-CoV were detected with two mAbs (1A5 and 2C5) targeting at a subdomain of S protein (aa 310-535), consistent with the previous report that this segment of S protein contains the major neutralizing domain. Some of these S-specific mAbs were able to recognize cleaved products of S protein in SARS-CoV-infected Vero E6 cells. None of the remaining five mAbs could recognize either of the recombinant S, N, M, or E antigens by ELISA. This study demonstrated that the inactivated SARS-CoV was able to preserve the immunogenicity of S protein including its major neutralizing domain. The relative ease with which these mAbs were generated against SARS-CoV virions further supports that subunit vaccination with S constructs may also be able to protect animals and perhaps humans. It is somewhat unexpected that no N-specific mAbs were identified albeit anti-N IgG was easily identified in SARS-CoV-infected patients. The availability of this panel of mAbs also provided potentially useful agents with applications in therapy, diagnosis, and basic research of SARS-CoV.
Figures


References
-
- Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., Weingartl H., Fernando L., Marszal P., Gren J., Nicolas B., Andonova M., Ranada F., Gubbins M.J., Ball T.B., Kitching P., Li Y., Kabani A., Plummer F. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods. 2004;120(1):87–96. - PMC - PubMed
-
- Che X.Y., Qiu L.W., Pan Y.X., Xu H., Hao W., Liao Z.Y., Mei Y.B., Zhang L.Y., Wan Z.Y., Yuan G.Y., Huang Z. Rapid and efficient preparation of monoclonal antibodies against SARS-associated coronavirus nucleocapsid protein by immunizing mice. Di Yi Jun Yi Da Xue Xue Bao. 2003;23(7):640–642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

