Effect of polymorphism on expression of the neuropeptide Y gene in inbred alcohol-preferring and -nonpreferring rats
- PMID: 15749341
- PMCID: PMC4455873
- DOI: 10.1016/j.neuroscience.2004.10.013
Effect of polymorphism on expression of the neuropeptide Y gene in inbred alcohol-preferring and -nonpreferring rats
Abstract
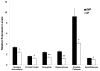
Using animal models of alcoholism, previous studies suggest that neuropeptide Y (NPY) may be implicated in alcohol preference and consumption due to its role in the modulation of feeding and anxiety. Quantitative trait loci (QTL) analysis previously identified an interval on rat chromosome 4 that is highly associated with alcohol preference and consumption using an F2 population derived from inbred alcohol-preferring (iP) and -nonpreferring (iNP) rats. NPY mapped to the peak of this QTL region and was prioritized as a candidate gene for alcohol-seeking behavior in the iP and iNP rats. In order to identify a potential mechanism for reduced NPY protein levels documented in the iP rat, genetic and molecular components that influence NPY expression were analyzed between iP and iNP rats. Comparing the iP rat to the iNP rat, quantitative real-time polymerase chain reaction detected significantly decreased levels of NPY mRNA expression in the iP rat in the six brain regions tested: nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate-putamen, and hypothalamus. In addition, the functional significance of three previously identified polymorphisms was assessed using in vitro expression analysis. The polymorphism defined by microsatellite marker D4Mit7 in iP rats reduced luciferase reporter gene expression in SK-N-SH neuroblastoma cells. These results suggest that differential expression of the NPY gene resulting from the D4mit7 marker polymorphism may contribute to reduced levels of NPY in discrete brain regions in the iP rats.
Figures


References
-
- Agarwal AK, Giacchetti G, Lavery G, Nikkila H, Palermo M, Ricketts M, McTernan C, Bianchi G, Manunta P, Strazzullo P, Mantero F, White PC, Stewart PM. CA-Repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension. 2000;36:187–194. - PubMed
-
- Arnold R, Maueler W, Bassili G, Lutz M, Burke L, Epplen TJ, Renkawitz R. The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA-DRB1(*)0401 gene. Gene. 2000;253:209–214. - PubMed
-
- Badia-Elder NE, Stewart RB, Powrozek TA, Roy KF, Murphy JM, Li TK. Effect of neuropeptide Y (NPY) on oral ethanol intake in Wistar, alcohol-preferring (P), and -nonpreferring (NP) rats. Alcohol Clin Exp Res. 2001;25:386–390. - PubMed
-
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. - PubMed
-
- Bream JH, Carrington M, O’Toole S, Dean M, Gerrard B, Shin HD, Kosack D, Modi W, Young HA, Smith MW. Polymorphisms of the human IFNG gene noncoding regions. Immunogenetics. 2000;51:50–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

