Auxin: regulation, action, and interaction
- PMID: 15749753
- PMCID: PMC4246732
- DOI: 10.1093/aob/mci083
Auxin: regulation, action, and interaction
Abstract
Background: The phytohormone auxin is critical for plant growth and orchestrates many developmental processes.
Scope: This review considers the complex array of mechanisms plants use to control auxin levels, the movement of auxin through the plant, the emerging view of auxin-signalling mechanisms, and several interactions between auxin and other phytohormones. Though many natural and synthetic compounds exhibit auxin-like activity in bioassays, indole-3-acetic acid (IAA) is recognized as the key auxin in most plants. IAA is synthesized both from tryptophan (Trp) using Trp-dependent pathways and from an indolic Trp precursor via Trp-independent pathways; none of these pathways is fully elucidated. Plants can also obtain IAA by beta-oxidation of indole-3-butyric acid (IBA), a second endogenous auxin, or by hydrolysing IAA conjugates, in which IAA is linked to amino acids, sugars or peptides. To permanently inactivate IAA, plants can employ conjugation and direct oxidation. Consistent with its definition as a hormone, IAA can be transported the length of the plant from the shoot to the root; this transport is necessary for normal development, and more localized transport is needed for tropic responses. Auxin signalling is mediated, at least in large part, by an SCFTIR1 E3 ubiquitin ligase complex that accelerates Aux/IAA repressor degradation in response to IAA, thereby altering gene expression. Two classes of auxin-induced genes encode negatively acting products (the Aux/IAA transcriptional repressors and GH3 family of IAA conjugating enzymes), suggesting that timely termination of the auxin signal is crucial. Auxin interaction with other hormone signals adds further challenges to understanding auxin response.
Conclusions: Nearly six decades after the structural elucidation of IAA, many aspects of auxin metabolism, transport and signalling are well established; however, more than a few fundamental questions and innumerable details remain unresolved.
Figures
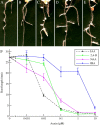
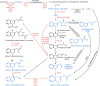


References
-
- Abel S, Nguyen MD, Chow W, Theologis A. 1995.ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana Journal of Biological Chemistry 270: 19093–19099. - PubMed
-
- Abel S, Nguyen MD, Theologis A. 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana Journal of Molecular Biology 251: 533–549. - PubMed
-
- Adham AR, Zolman BK, Millius A, Bartel B. 2005. Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant Journal 41: 859–874. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

