Timing of the maternal-to-zygotic transition during early seed development in maize
- PMID: 15749756
- PMCID: PMC1087986
- DOI: 10.1105/tpc.104.029819
Timing of the maternal-to-zygotic transition during early seed development in maize
Abstract
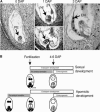
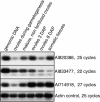
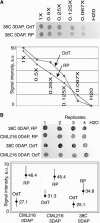
In animals, early embryonic development is largely dependent on maternal transcripts synthesized during gametogenesis. Recent data in plants also suggest maternal control over early seed development, but the actual timing of zygotic genome activation is unclear. Here, we analyzed the timing of the maternal-to-zygotic transition during early Zea mays seed development. We show that for 16 genes expressed during early seed development, only maternally inherited alleles are detected during 3 d after fertilization in both the embryo and the endosperm. Microarray analyses of precocious embryonic development in apomictic hybrids between maize and its wild relative, Tripsacum, demonstrate that early embryo development occurs without significant quantitative changes to the transcript population in the ovule before fertilization. Precocious embryo development is also correlated with a higher proportion of polyadenylated mRNA in the ovules. Our data suggest that the maternal-to-zygotic transition occurs several days after fertilization. By contrast, novel transcription accompanies early endosperm development, indicating that different mechanisms are involved in the initiation of endosperm and embryo development.
Figures


References
-
- Baird, D., Johnstone, P., and Wilson, T. (2004). Normalization of microarray data using a spatial mixed model analysis which includes splines. Bioinformatics 22, 3196–3205. - PubMed
-
- Baroux, C., Blanvillain, R., and Gallois, P. (2001). Paternally inherited transgenes are down-regulated but retain low activity during early embryogenesis in Arabidopsis. FEBS Lett. 509, 11–16. - PubMed
-
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
-
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E., and Fischer, R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

