Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard
- PMID: 15750058
- PMCID: PMC1081233
- DOI: 10.1128/JCM.43.3.1045-1050.2005
Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard
Abstract
The PulseNet National Database, established by the Centers for Disease Control and Prevention in 1996, consists of pulsed-field gel electrophoresis (PFGE) patterns obtained from isolates of food-borne pathogens (currently Escherichia coli O157:H7, Salmonella, Shigella, and Listeria) and textual information about the isolates. Electronic images and accompanying text are submitted from over 60 U.S. public health and food regulatory agency laboratories. The PFGE patterns are generated according to highly standardized PFGE protocols. Normalization and accurate comparison of gel images require the use of a well-characterized size standard in at least three lanes of each gel. Originally, a well-characterized strain of each organism was chosen as the reference standard for that particular database. The increasing number of databases, difficulty in identifying an organism-specific standard for each database, the increased range of band sizes generated by the use of additional restriction endonucleases, and the maintenance of many different organism-specific strains encouraged us to search for a more versatile and universal DNA size marker. A Salmonella serotype Braenderup strain (H9812) was chosen as the universal size standard. This strain was subjected to rigorous testing in our laboratories to ensure that it met the desired criteria, including coverage of a wide range of DNA fragment sizes, even distribution of bands, and stability of the PFGE pattern. The strategy used to convert and compare data generated by the new and old reference standards is described.
Figures

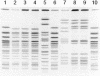



References
-
- Applied Maths BVBA. 2002. BioNumerics version 3.0 manual. Applied Maths BVBA, Sint-Martens-Latem, Belgium.
-
- Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. - PubMed
-
- Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. - PubMed
-
- Branchini, M. L. 1993. Application of genomic DNA subtyping by pulsed field gel electrophoresis and restriction enzyme analysis of plasmid DNA to characterize methicillin-resistant Staphylococcus aureus from two nosocomial outbreaks. J. Hosp. Infect. 25:109-116. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

