Predominance of porcine rotavirus G9 in Japanese piglets with diarrhea: close relationship of their VP7 genes with those of recent human G9 strains
- PMID: 15750112
- PMCID: PMC1081228
- DOI: 10.1128/JCM.43.3.1377-1384.2005
Predominance of porcine rotavirus G9 in Japanese piglets with diarrhea: close relationship of their VP7 genes with those of recent human G9 strains
Abstract
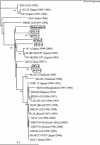
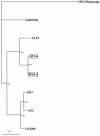
Type G9 of group A rotavirus (GAR) was shown to be predominant in a survey of VP7 (G) and VP4 (P) genotypes among porcine GARs associated with outbreaks of diarrhea in young pigs in Japan between 2000 and 2002. Comparison of the G9 VP7 gene sequences showed that the porcine G9 strains were more closely related to human G9 strains reemerging globally since the mid-1990s than to those from the mid-1980s. The VP7 gene sequences of porcine G9 strains from different farms were divergent (6.1 to 7.2% difference in nucleotides), suggesting that these G9 VP7 genes were not the result of recent introduction into the porcine population. Regarding the P genotype specificities of porcine G9 strains, while the majority of strains were close to unusual porcine P types (P[13] and P[23]), two strains were of the P[6] type, which has closer sequence identity with the human AU19 strain than with the porcine Gottfried strain. These unexpected results suggest that G9 GARs in the porcine population have spread more widely than previously thought and that the VP7 genes of porcine G9 strains and those of some human G9 strains detected recently may have a common progenitor.
Figures


References
-
- Burke, B., M. A. McCrae, and U. Desselberger. 1994. Sequence analysis of two porcine rotaviruses differing in growth in vitro and in pathogenicity: distinct VP4 sequences and conservation of NS53, VP6 and VP7 genes. J. Gen. Virol. 75:2205-2212. - PubMed
-
- Fitzgerald, T. A., M. Munoz, A. R. Wood, and D. R. Snodgrass. 1995. Serological and genomic characterisation of group A rotaviruses from lambs. Arch. Virol. 140:1541-1548. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

