Basal immunoglobulin signaling actively maintains developmental stage in immature B cells
- PMID: 15752064
- PMCID: PMC1059451
- DOI: 10.1371/journal.pbio.0030082
Basal immunoglobulin signaling actively maintains developmental stage in immature B cells
Abstract
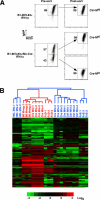
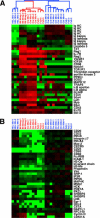
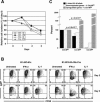
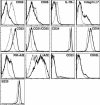
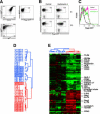
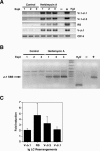
In developing B lymphocytes, a successful V(D)J heavy chain (HC) immunoglobulin (Ig) rearrangement establishes HC allelic exclusion and signals pro-B cells to advance in development to the pre-B stage. A subsequent functional light chain (LC) rearrangement then results in the surface expression of IgM at the immature B cell stage. Here we show that interruption of basal IgM signaling in immature B cells, either by the inducible deletion of surface Ig via Cre-mediated excision or by incubating cells with the tyrosine kinase inhibitor herbimycin A or the phosphatidylinositol 3-kinase inhibitor wortmannin, led to a striking "back-differentiation" of cells to an earlier stage in B cell development, characterized by the expression of pro-B cell genes. Cells undergoing this reversal in development also showed evidence of new LC gene rearrangements, suggesting an important role for basal Ig signaling in the maintenance of LC allelic exclusion. These studies identify a previously unappreciated level of plasticity in the B cell developmental program, and have important implications for our understanding of central tolerance mechanisms.
Figures

References
-
- Alt F, Blackwell K, Yancopoulos G. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. - PubMed
-
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. - PubMed
-
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

