Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation
- PMID: 15752426
- PMCID: PMC1079855
- DOI: 10.1186/1471-2180-5-8
Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation
Abstract
Background: Several strains of bacteria have sequenced and annotated genomes, which have been used in conjunction with biochemical and physiological data to reconstruct genome-scale metabolic networks. Such reconstruction amounts to a two-dimensional annotation of the genome. These networks have been analyzed with a constraint-based formalism and a variety of biologically meaningful results have emerged. Staphylococcus aureus is a pathogenic bacterium that has evolved resistance to many antibiotics, representing a significant health care concern. We present the first manually curated elementally and charge balanced genome-scale reconstruction and model of S. aureus' metabolic networks and compute some of its properties.
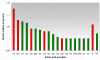
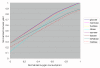
Results: We reconstructed a genome-scale metabolic network of S. aureus strain N315. This reconstruction, termed iSB619, consists of 619 genes that catalyze 640 metabolic reactions. For 91% of the reactions, open reading frames are explicitly linked to proteins and to the reaction. All but three of the metabolic reactions are both charge and elementally balanced. The reaction list is the most complete to date for this pathogen. When the capabilities of the reconstructed network were analyzed in the context of maximal growth, we formed hypotheses regarding growth requirements, the efficiency of growth on different carbon sources, and potential drug targets. These hypotheses can be tested experimentally and the data gathered can be used to improve subsequent versions of the reconstruction.
Conclusion: iSB619 represents comprehensive biochemically and genetically structured information about the metabolism of S. aureus to date. The reconstructed metabolic network can be used to predict cellular phenotypes and thus advance our understanding of a troublesome pathogen.
Figures
References
-
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

